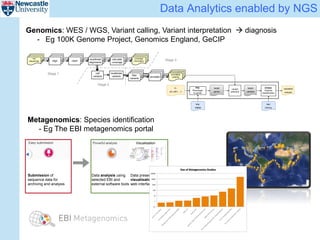
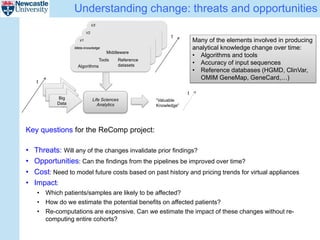
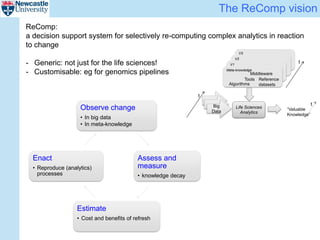
The ReComp project aims to enable selective re-computation of genomic pipelines in response to changes, focusing on improving the efficiency and accuracy of genomic data analysis processes while considering the impact on patients. It seeks to address challenges such as learning from past analytics, ensuring reproducibility, and optimizing software infrastructure, all through effective metadata management. Funded by EPSRC, the project builds on previous initiatives to facilitate reliable genetic testing and workflow-based processing of next-generation sequencing (NGS) pipelines.