SNP Genotyping Technologies
•
18 likes•12,437 views
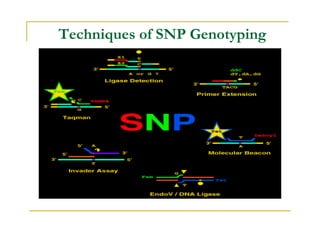
Techniques of SNP Genotyping can be summarized as follows: There are several techniques for genotyping SNPs including hybridization methods, enzyme-based methods, and other methods based on physical properties of DNA. Popular hybridization methods include DASH, molecular beacons, and gene chip arrays. Common enzyme-based techniques are RFLP, Invader assay, and oligonucleotide ligation assay. Other physical property-based methods include SSCP, TGGE, and pyrosequencing. Each method has its own pros and cons related to factors like speed, cost, and accuracy. Choosing the appropriate SNP genotyping technique depends on the number of SNPs needed to be analyzed and sample size.
Report
Share
Report
Share
Download to read offline
Recommended
Recommended
More Related Content
What's hot
What's hot (20)
Next generation sequencing technologies for crop improvement

Next generation sequencing technologies for crop improvement
Transcriptomics: A time efficient tool for crop improvement

Transcriptomics: A time efficient tool for crop improvement
Similar to SNP Genotyping Technologies
Please note: This presentation accompanies a recorded webinar at:
https://www1.gotomeeting.com/register/347794241
Biomarkers for studying gene regulation and cell function can be efficiently analyzed by multiplexed methods. Dr. Jim Lazar from OriGene Technologies will provide an overview of four different but related detection technologies that can be used to analyze genetic variants, microRNA expression, transcription factor binding, and protein expression on the Luminex xMAP platform. OriGene’s broad panel of assays and tools for discovery, analysis and validation of multiple classes of important biomarkers will allow researcher to develop more accurate descriptions of biologically complex systems.Multiplex Assays for Studying Gene Regulation and Cell Function

Multiplex Assays for Studying Gene Regulation and Cell FunctionMiraiBio Group of Hitachi Solutions America, Ltd.
Similar to SNP Genotyping Technologies (20)
Fruit breedomics workshop wp6 from marker assisted breeding to genomics assis...

Fruit breedomics workshop wp6 from marker assisted breeding to genomics assis...
2007. stephen chanock. technologic issues in gwas and follow up studies

2007. stephen chanock. technologic issues in gwas and follow up studies
Multiplex Assays for Studying Gene Regulation and Cell Function

Multiplex Assays for Studying Gene Regulation and Cell Function
DNA MARKERS 2023 DNA FINGERPRINTING TYPE OF METHODS OF DNA FINGERPRINTING

DNA MARKERS 2023 DNA FINGERPRINTING TYPE OF METHODS OF DNA FINGERPRINTING
Understanding mechanisms underlying human gene expression variation with RNA ...

Understanding mechanisms underlying human gene expression variation with RNA ...
Recently uploaded
Recently uploaded (20)
ICT role in 21st century education and its challenges

ICT role in 21st century education and its challenges
Apidays New York 2024 - Accelerating FinTech Innovation by Vasa Krishnan, Fin...

Apidays New York 2024 - Accelerating FinTech Innovation by Vasa Krishnan, Fin...
Connector Corner: Accelerate revenue generation using UiPath API-centric busi...

Connector Corner: Accelerate revenue generation using UiPath API-centric busi...
TrustArc Webinar - Stay Ahead of US State Data Privacy Law Developments

TrustArc Webinar - Stay Ahead of US State Data Privacy Law Developments
AWS Community Day CPH - Three problems of Terraform

AWS Community Day CPH - Three problems of Terraform
Strategies for Unlocking Knowledge Management in Microsoft 365 in the Copilot...

Strategies for Unlocking Knowledge Management in Microsoft 365 in the Copilot...
TrustArc Webinar - Unlock the Power of AI-Driven Data Discovery

TrustArc Webinar - Unlock the Power of AI-Driven Data Discovery
Navi Mumbai Call Girls 🥰 8617370543 Service Offer VIP Hot Model

Navi Mumbai Call Girls 🥰 8617370543 Service Offer VIP Hot Model
Mastering MySQL Database Architecture: Deep Dive into MySQL Shell and MySQL R...

Mastering MySQL Database Architecture: Deep Dive into MySQL Shell and MySQL R...
How to Troubleshoot Apps for the Modern Connected Worker

How to Troubleshoot Apps for the Modern Connected Worker
Strategize a Smooth Tenant-to-tenant Migration and Copilot Takeoff

Strategize a Smooth Tenant-to-tenant Migration and Copilot Takeoff
Axa Assurance Maroc - Insurer Innovation Award 2024

Axa Assurance Maroc - Insurer Innovation Award 2024
"I see eyes in my soup": How Delivery Hero implemented the safety system for ...

"I see eyes in my soup": How Delivery Hero implemented the safety system for ...
SNP Genotyping Technologies
- 1. Techniques of SNP Genotyping
- 2. SNPs Single Nucleotide Polymorphisms Single Nucleotide variation at a specific locationSingle Nucleotide variation at a specific location in the genome SNP: Commonly biallelic C T Commonly found
- 3. Regulatory sites Coding Regions Exons Introns Non coding region 3’ end5’ end SNP: Alterations in protein structure Non coding region Regulatory sites (Promoter) Introns Non coding region SNP: Transcription rate changes Encoded protein production changes High frequency Evolutionarily stable Unique in particular geographical or ethnic group important markers for comparative and Evolutionary genomics studies
- 4. Popularity of different Polymorphism studies
- 6. How many SNPs are there in Humans today? Human Mutation rate is ~2.5 x 10-8 mutations/site/gen ~150 mutations/diploidgenome/generation 6.8 billion people in the world =1,020,000,000,000 mutations in the=1,020,000,000,000 mutations in the world today. With 3 billion nucleotides = each nucleotide in the world today is mutated 340 times.
- 7. What can SNPs tell you? Linkage Disequilibrium Recombination Association Studies Demographic eventsDemographic events
- 8. Linkage Disequilibrium The non-random association between alleles in a population. No LD 2 SNPs 4 Haplotypes High LD 2 SNPs 2 Haplotyps
- 9. Association studies An association between a genetic variant and a phenotype. Two groups: Disease Group Control (random pop) Look for a variant which is in high freq in your disease group in respect to the control. Cystic Fibrosis was the first successful association study, based on RFLPs. (Kerem et al Science 1989) SNP at Promoter (-169C- susceptible/T non susceptible) region of FCRL3 for Rheumatoid arthritis in Japanese population (Kochi et al., Nat Genet. 2005 Jun; 37(6):652.)
- 10. SNP typing Techniques Hybridization Methods Enzyme based Methods Other methods based on physical Properties of DNA 1. DASH 2. Molecular beacons 1. RFLP 2. Invader Assay 1. SSCP 2. TGGE Some commercial techniques 3. Primer Extension 4. Oligonucleotide ligation assay 5. Taqman assay 2. TGGE 1. Gene chip array- Affymetrix 2. Bead array- Illumina 3. Pyrosequencing – Illumina
- 11. DASH PCR is performed using one of the primers contains a 5’ biotin. Resulting products immobilized by a streptavidin-coated microtiter plate. The non-biotynylated strand is removed by NaOH solution. Hybridized with an oligonucleotide probe, (hybridization buffer containing a fluorescent double-strand-specificbuffer containing a fluorescent double-strand-specific intercalating dye). The sample is heated slowly from room temperature to above denaturing temperature while continually monitoring fluorescence. By plotting the negative derivative (slope of the fluorescence Vs temp.), denaturation points are clearly seen as peaks. Peak temperature values can be used for final allele determination.
- 12. DASH
- 14. Genomic DNA PCR amplified SNP region RE Digestion SNP-RFLPRE Digestion Gel electrophoresis SNP genotyping SNP-RFLP Genotyping
- 15. Invader assay 3’ 5’3’ 5’
- 17. Taqman assay
- 18. Taqman assay
- 19. Primer extension (CPE- Mass analysis) Mass analysis MALDI-TOF MS Commercial SNP assays: Pin pint assay Mass EXTEND SPC-SBE GOOD assay
- 20. CPE- Fluorescence analysis Commercial SNP assays: SNaPshot approach (Applied Biosystems) SNP stream assay (Orchid Biosciences) Fluorescence analysis
- 21. Primer extension (SPE- Mass analysis) Tag-It approach (Tm Bioscience corp, Canada)
- 22. SSCP Genomic DNA PCR amplification Denatured by heat/Denatured by heat/ formaldehyde Run on non- denaturing electrophoresis gel
- 23. Pros & cons PROS Rapid Technically simple Inexpensive Uses commonly available equipment Detects 60-95% of mutations in short DNA strandsDetects 60-95% of mutations in short DNA strands CONS Need to sequence DNA to identify specific mutation ssDNA conformation is highly dependent on temperature and it is not generally apparent what the ideal temperature is. Sensitivity drops when sequences longer than 400 bp are used (Costabile et al. 2006).
- 24. C G G C C G G GG C C A A T Normal DNA Target DNA Denaturing followed by annealing Homoduplexes Homoduplexes Heteroduplexes C G C A T T Homoduplexes Homoduplexes Heteroduplexes TGE TGE TGE
- 26. Invader assay- Third wave technologies
- 27. Genechip array- Affymetrix Allele-specific probes consisting of 25-mer oligonucleotides are synthesized in an orderedsynthesized in an ordered fashion to form a probe array
- 28. Pyrosequencing Mix genomic DNA with PCR reagents & thermocyle Purify biotinylated PCR products using Streptavidin-Sepharose Anneal extension primer to single stranded biotinylated PCR product Pyrosequencing reaction This methodology is good for small numbers of SNPssmall numbers of SNPs (less than 15) and on small sample sizes (1-1000)
- 32. Thank You … !Thank You … !