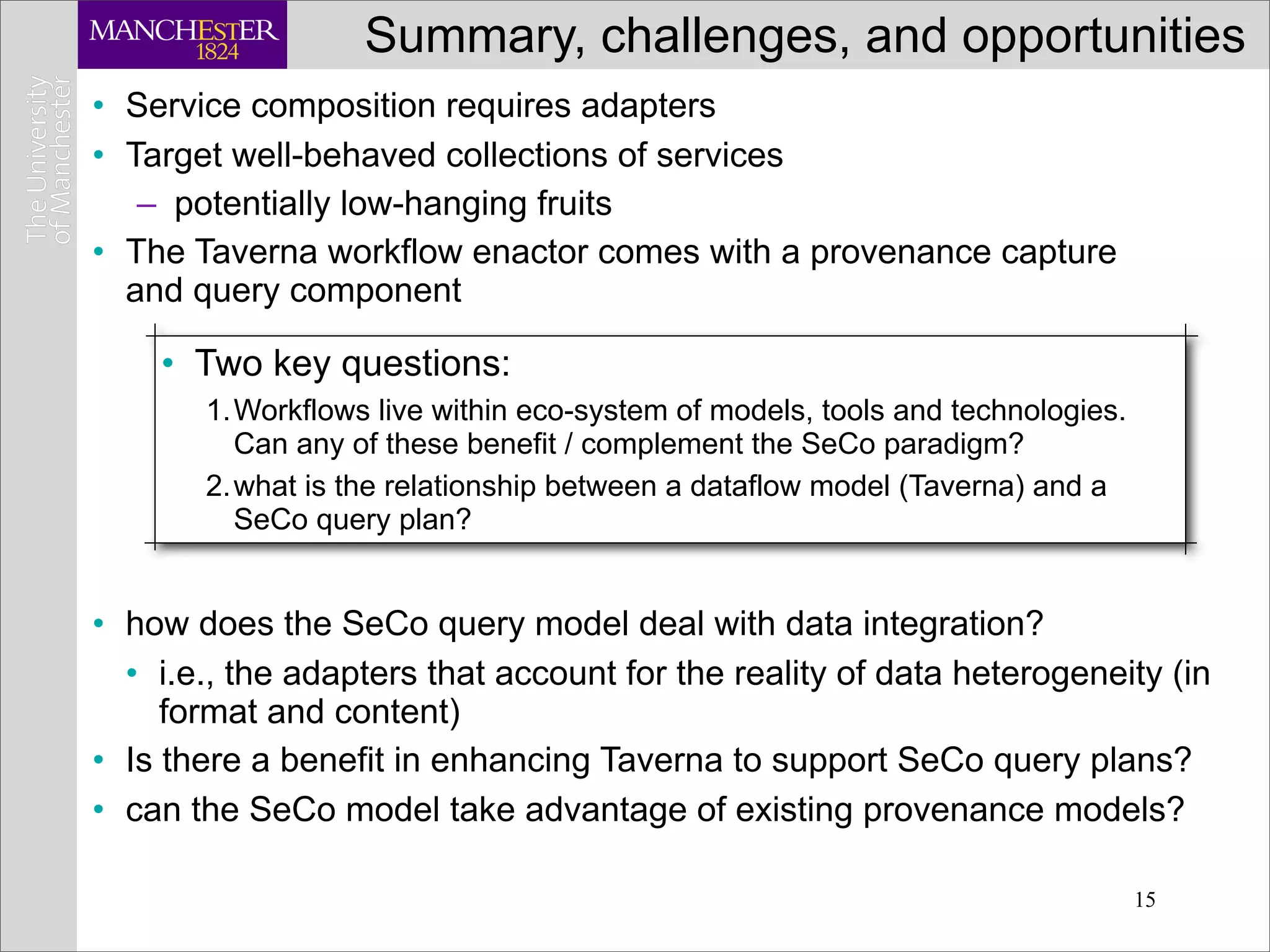
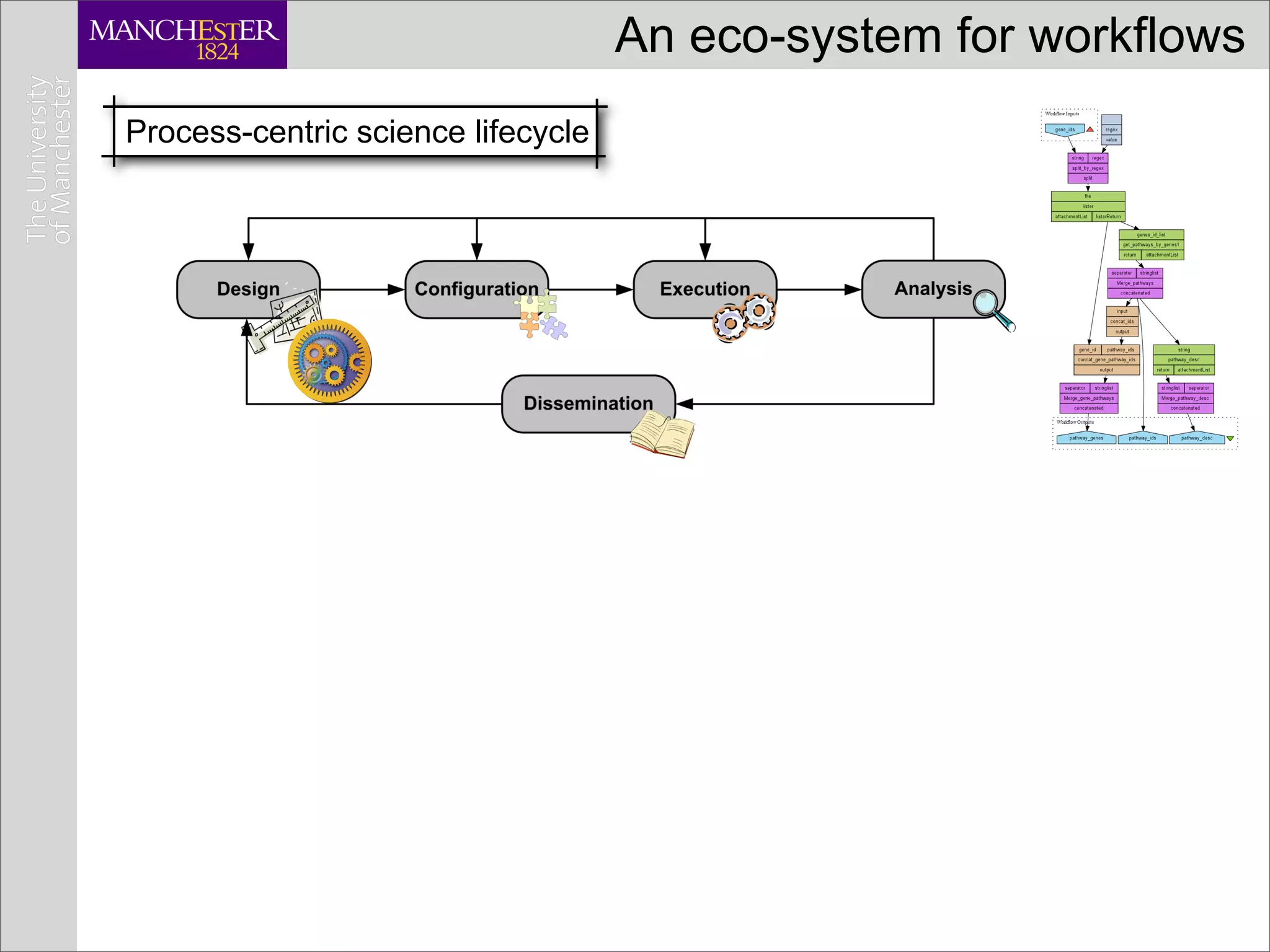
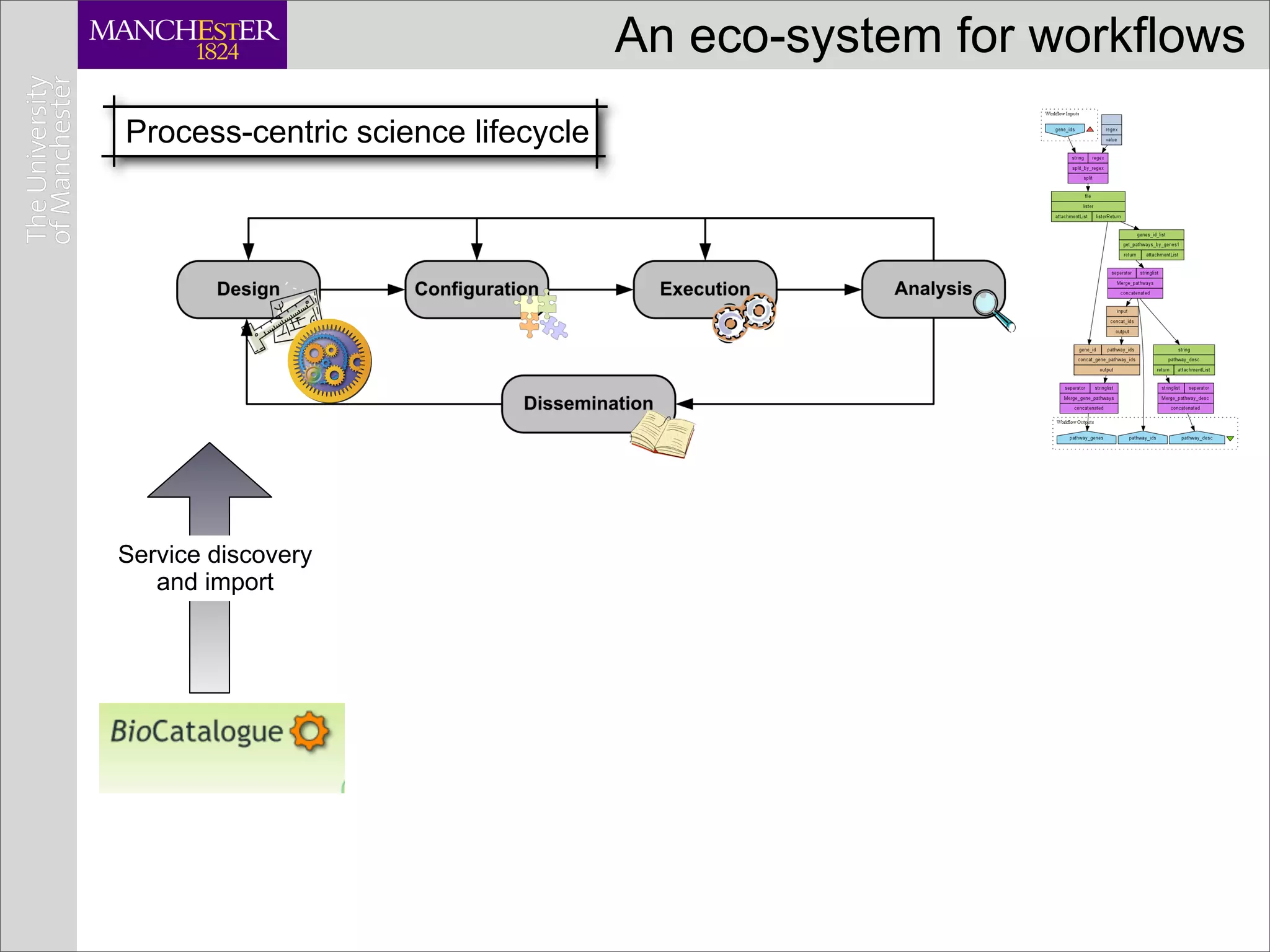
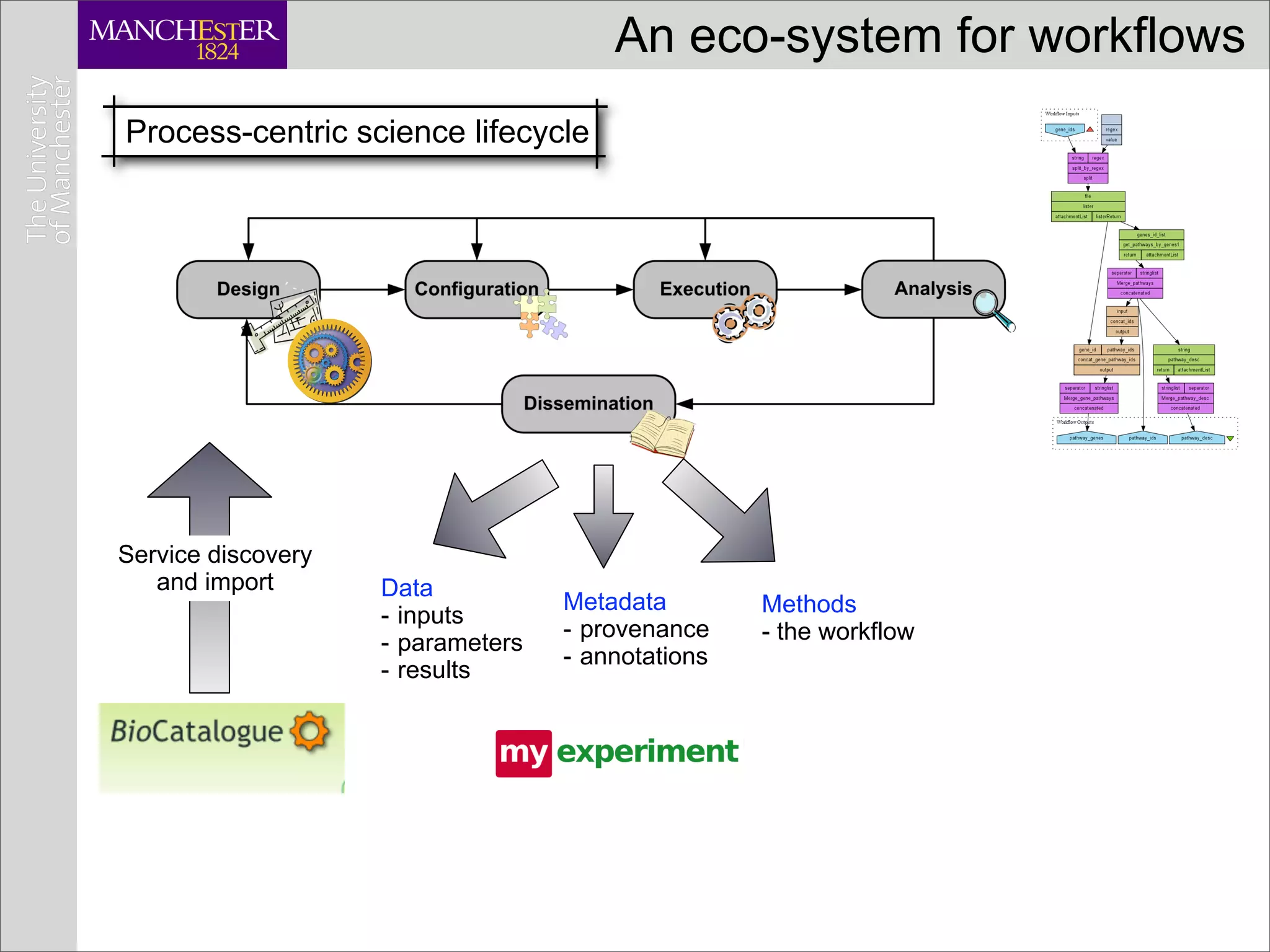
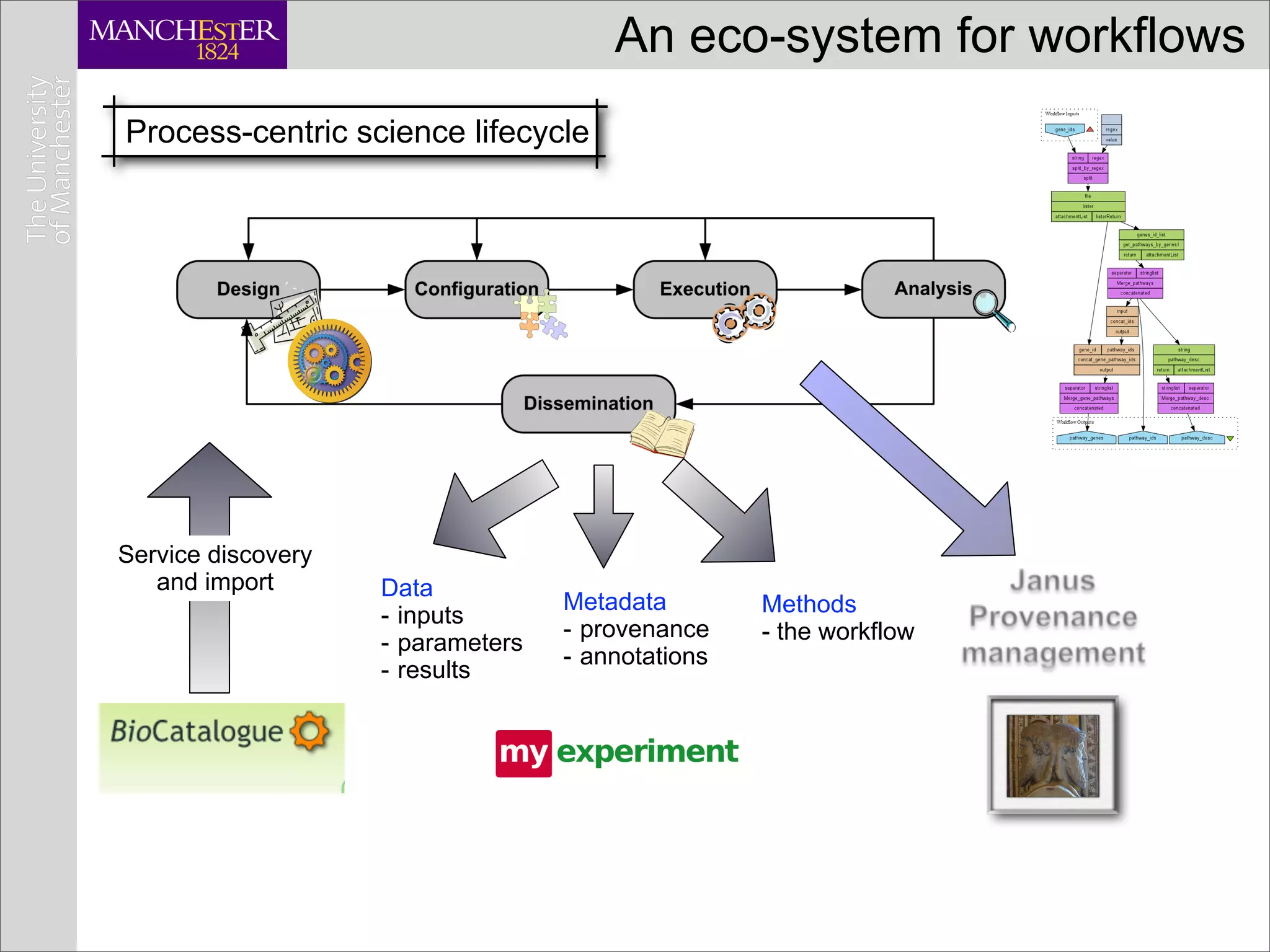
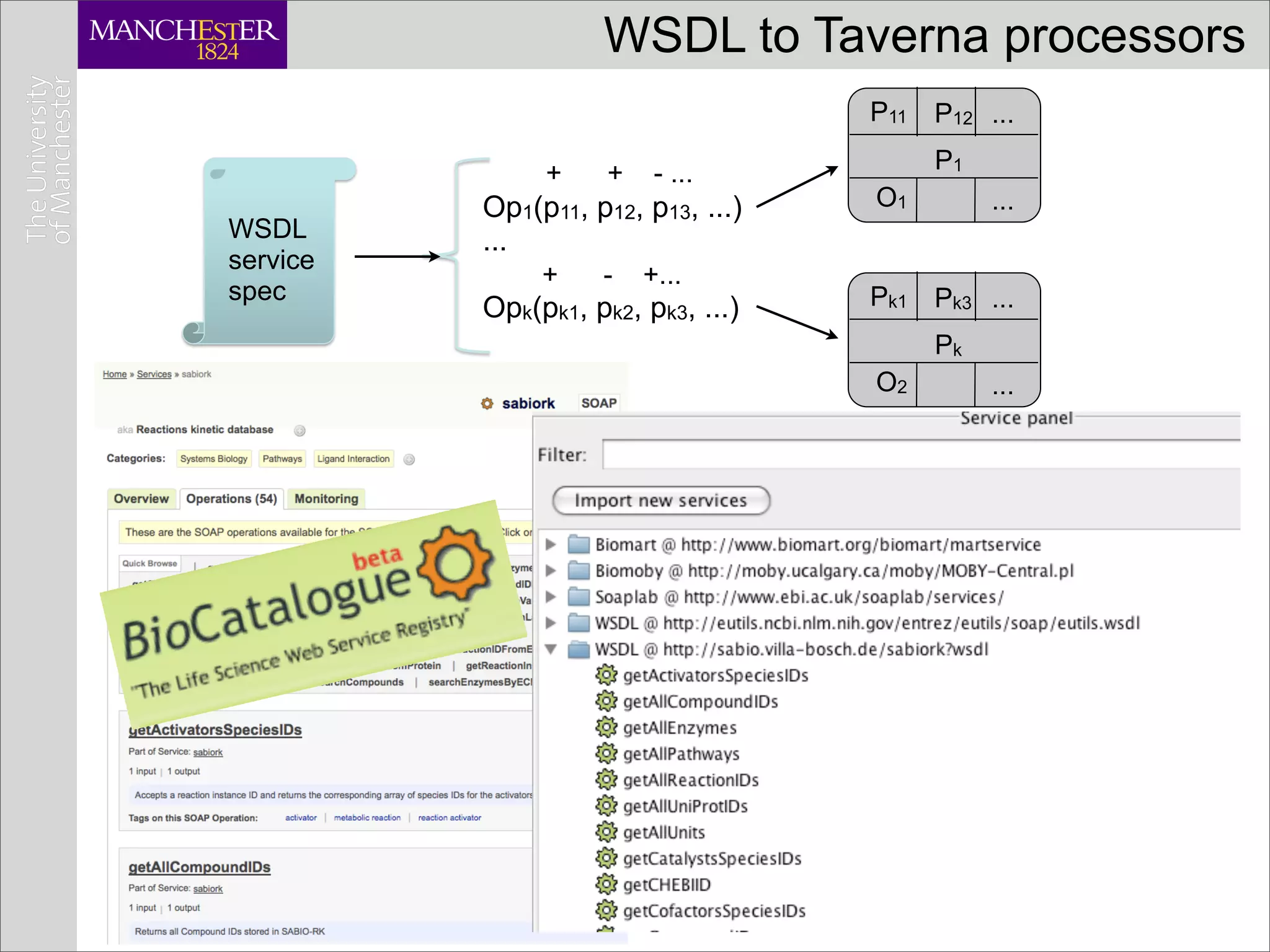
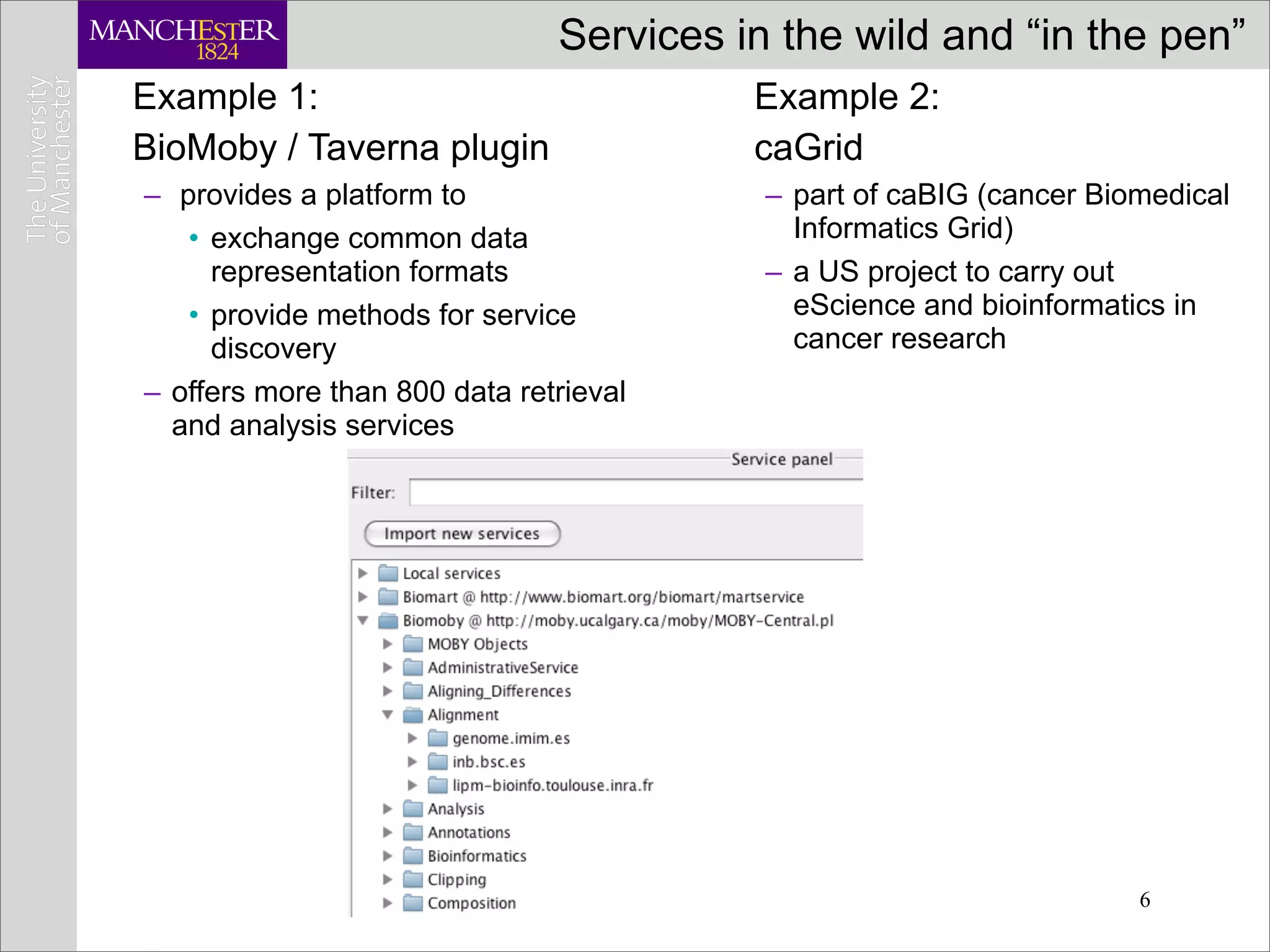
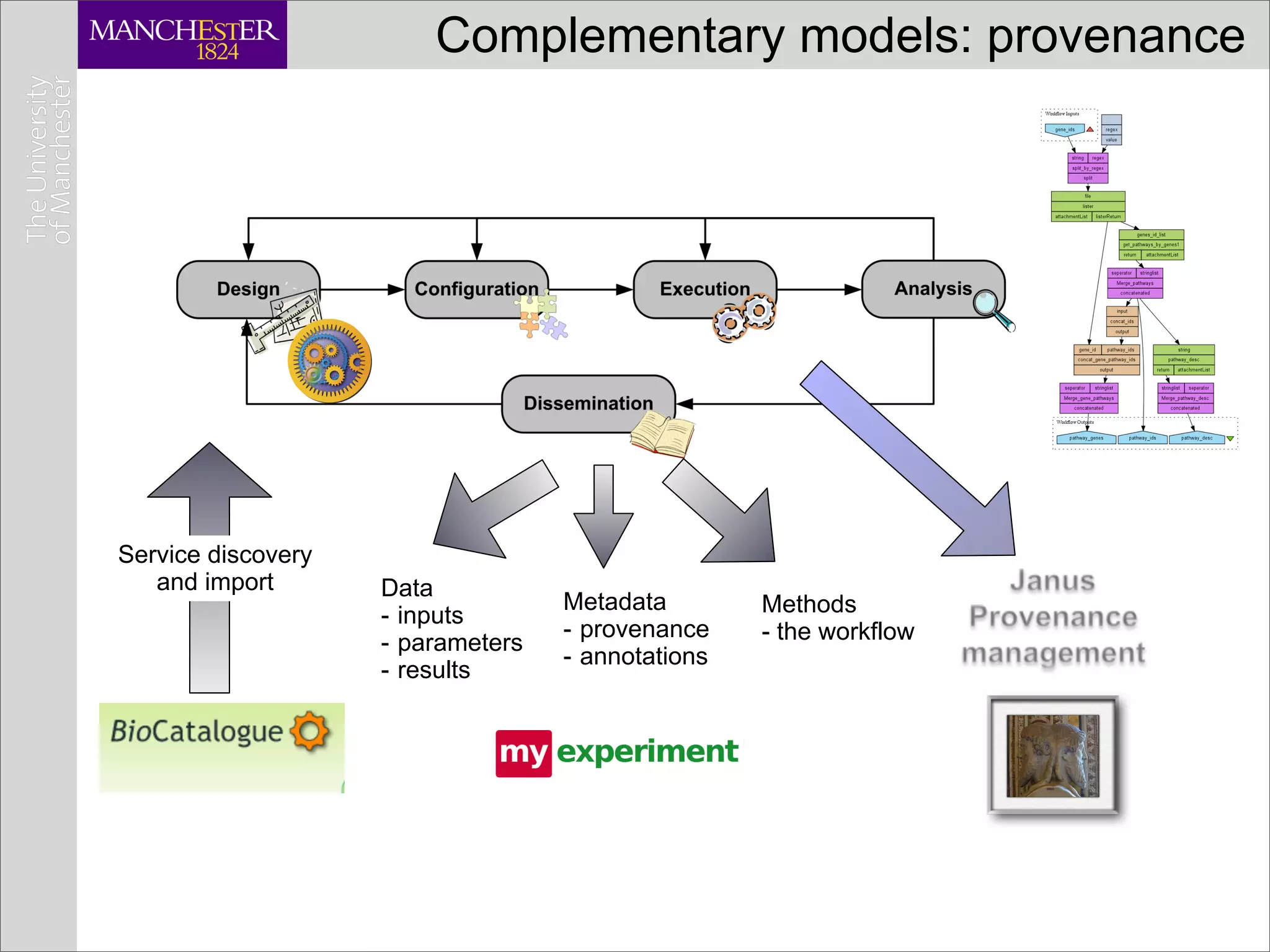
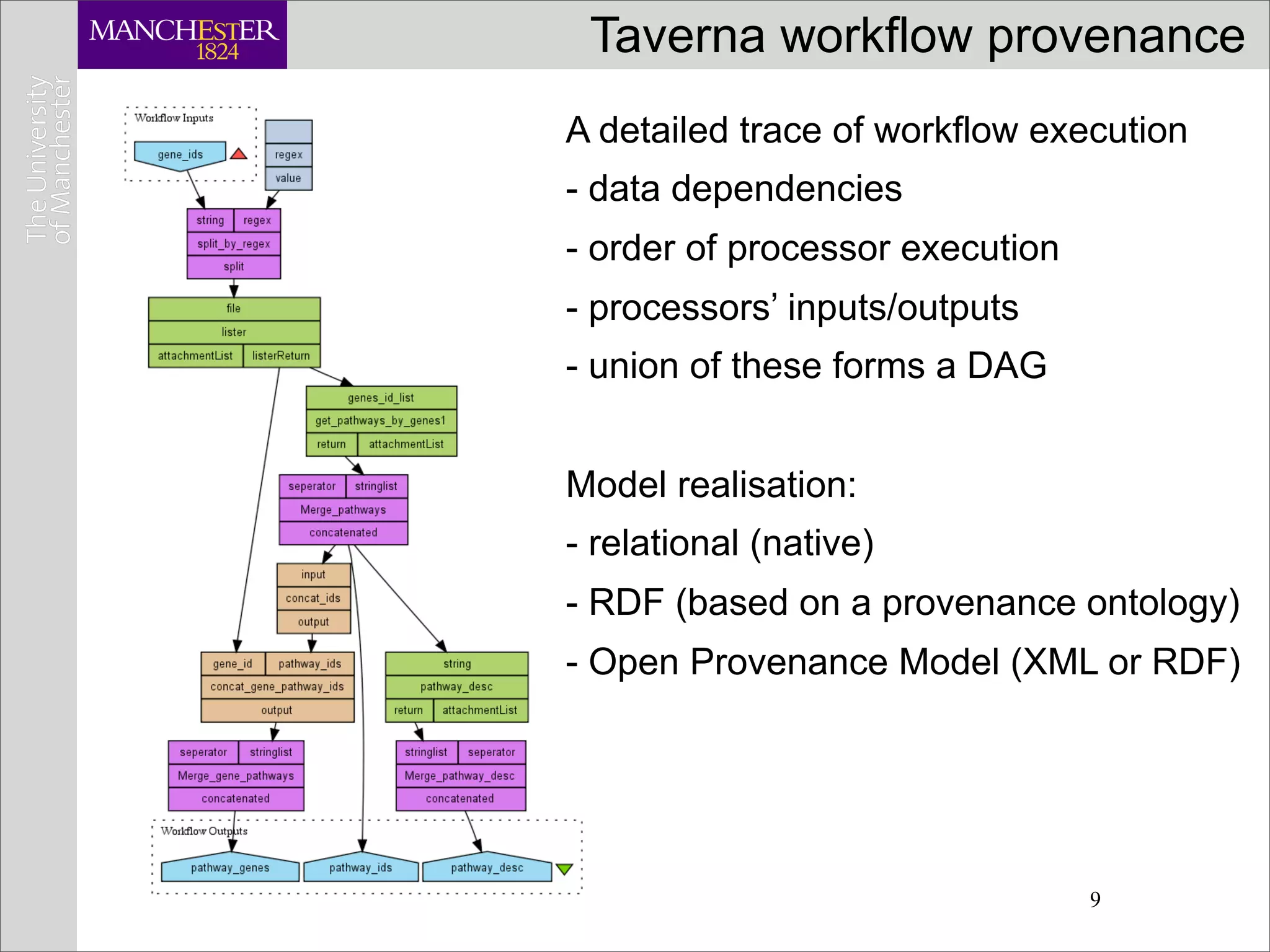
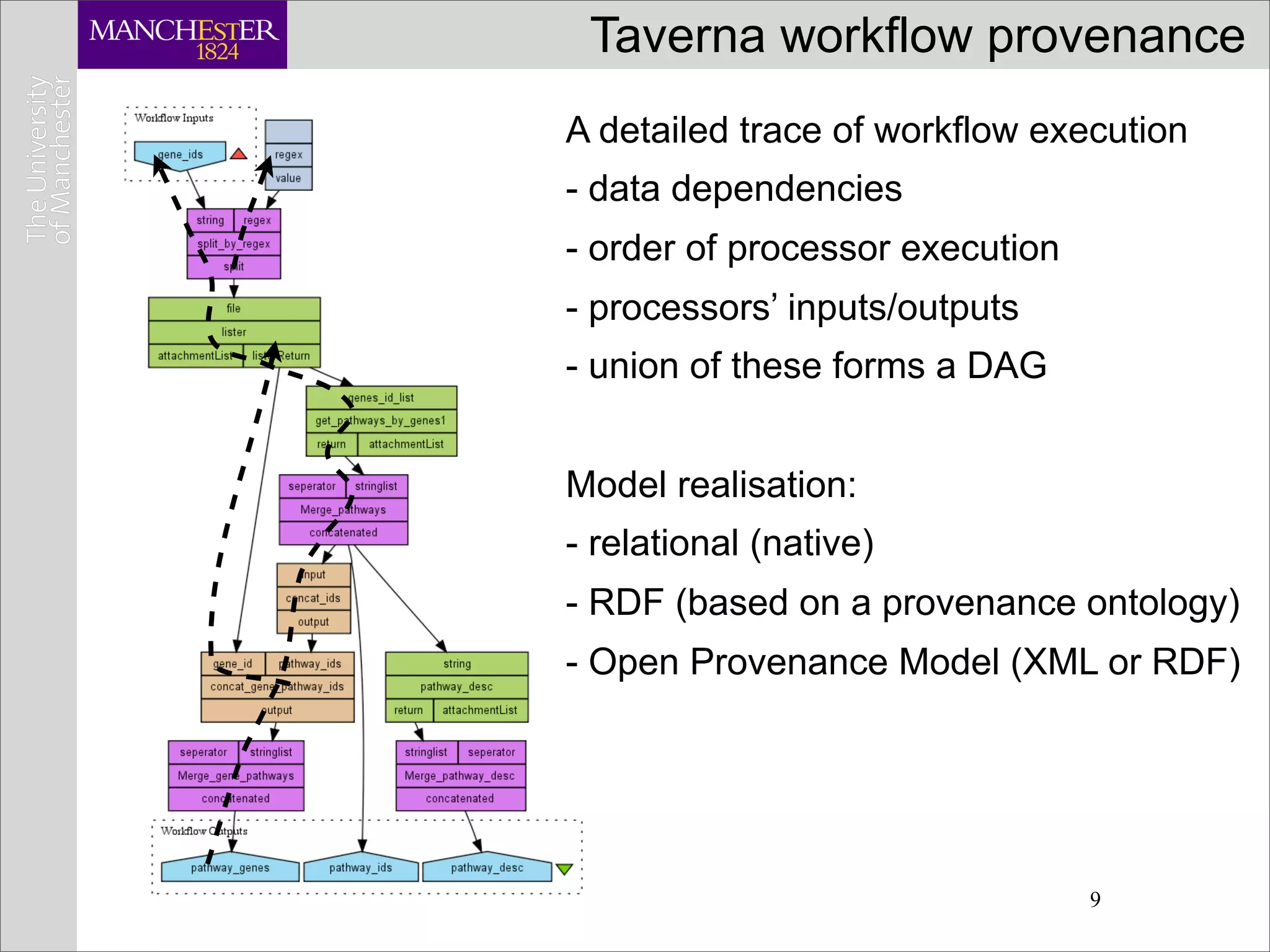
This document discusses workflows in the context of an ecosystem of models, tools, and technologies that can benefit or complement the SeCo paradigm. It focuses on some elements of a workflow's lifecycle, including importing services, benefits of domain-specific service collections, and collecting and querying provenance traces. Well-curated domain-specific service collections have advantages like easier discoverability and built-in interoperability compared to general collections. Provenance metadata captured during workflow runs can provide important context about the data and process.


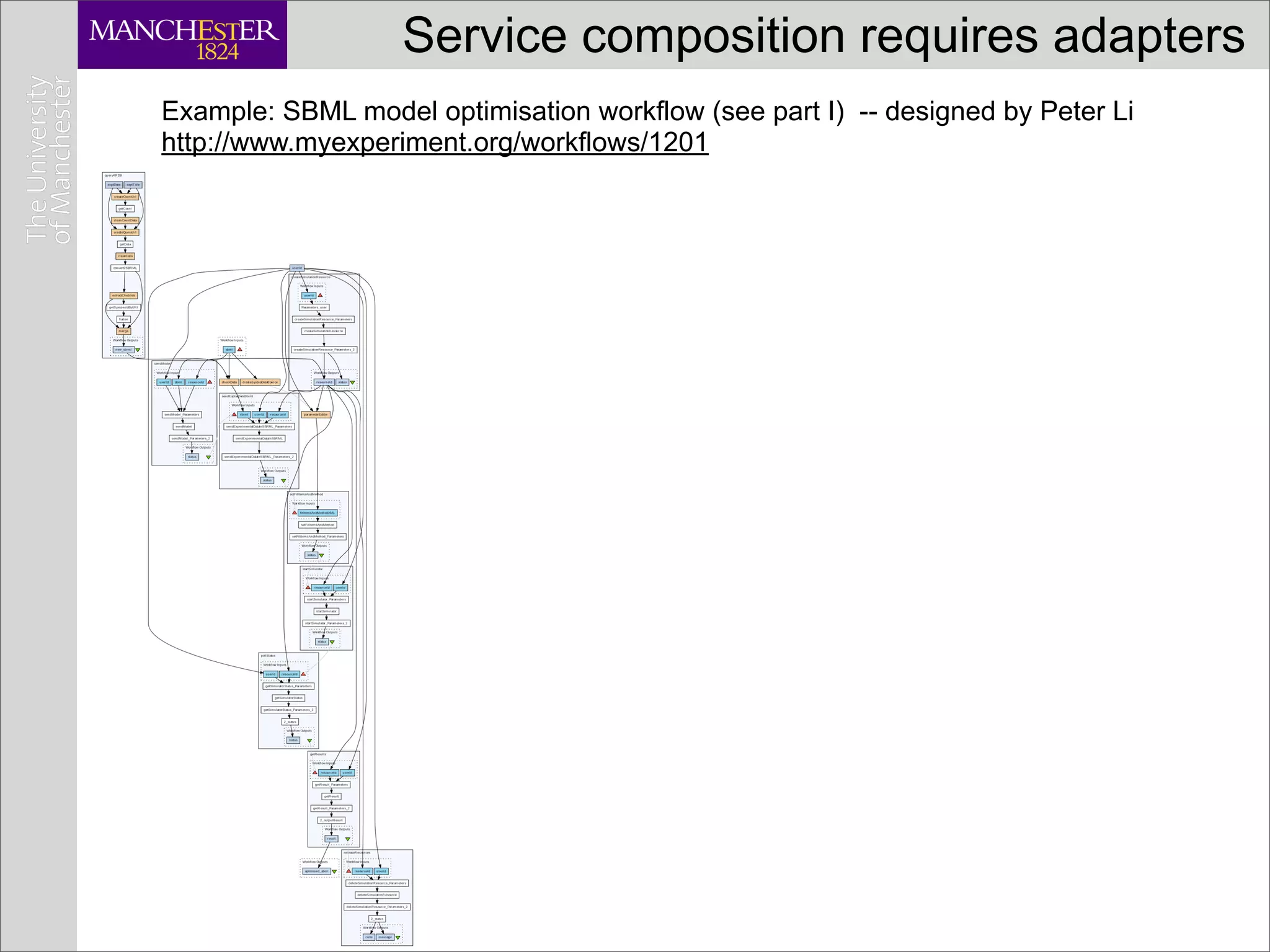
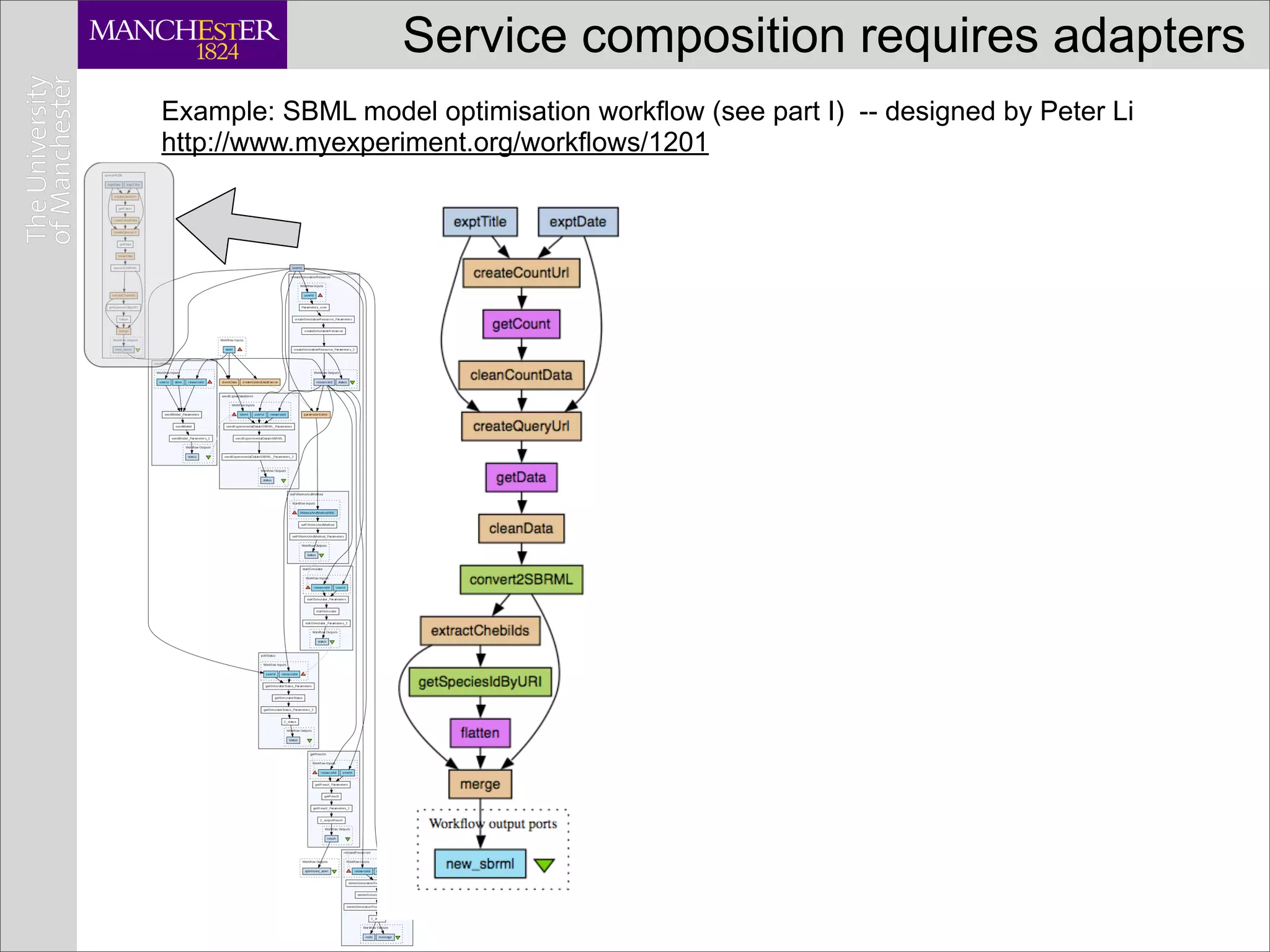
![Service composition requires adapters
Example: SBML model optimisation workflow (see part I) -- designed by Peter Li
http://www.myexperiment.org/workflows/1201
String[] lines = inStr.split("n");
StringBuffer sb = new StringBuffer();
for(i = 1; i < lines.length -1; i++)
{
String str = lines[i];
str = str.replaceAll("<result>", "");
str = str.replaceAll("</result>", "");
sb.append(str.trim() + "n");
}
String outStr = sb.toString();
Url -> content (built-in shell script)
import java.util.regex.Pattern;
import java.util.regex.Matcher;
sb = new StringBuffer();
p = "CHEBI:[0-9]+";
Pattern pattern = Pattern.compile(p);
Matcher matcher = pattern.matcher(sbrml);
while (matcher.find())
{
sb.append("urn:miriam:obo.chebi:" + matcher.group() + ",");
}
String out = sb.toString();
//Clean up
if(out.endsWith(","))
out = out.substring(0, out.length()-1);
chebiIds = out.split(",");](https://image.slidesharecdn.com/seco-workshop-paolo-100622043916-phpapp02/75/Invited-talk-Second-Search-Computing-workshop-13-2048.jpg)




![Querying Taverna provenance
provenance access Provenance
API (Java) capture
<select>
<workflow name="lymphoma"> Query
<processor name="fetch">
<outputPort name="value"
index="[1,2]" />
</processor>
...
</select>
<focus> Provenance
<workflow name="exprAnalysis"> lineage DB
<processor name="p1" />
</workflow> query
...
</focus>
processor
Native query answer [1]
(Java objects)
RDF query answer
[2] (Janus ontology)
Tupelo
OPM graph
(RDF/XML) OPM RDF
OPM graph graph
[3]
manager manager
11](https://image.slidesharecdn.com/seco-workshop-paolo-100622043916-phpapp02/75/Invited-talk-Second-Search-Computing-workshop-25-2048.jpg)
![Taverna computational model (very briefly)
[ [ mmu:26416, ... ],
[ mmu:328788, ... ] ]
• Collection processing
• Simple type system
• no record / tuple structure
• data driven computation
• with optional processor synchronisation
• parallel processor activation
• greedy (no scheduler)
[ path:mmu04010 MAPK signaling,
path:mmu04370 VEGF signaling ]
[ [ path:mmu04210 Apoptosis, path:mmu04010 MAPK signaling, ...],
[ path:mmu04010 MAPK signaling , path:mmu04620 Toll-like receptor, ...] ]
EDBT, Lausanne, March 2010](https://image.slidesharecdn.com/seco-workshop-paolo-100622043916-phpapp02/75/Invited-talk-Second-Search-Computing-workshop-27-2048.jpg)
![Parallelism in Taverna
• intra-processor: implicit iteration over collections
• inter-processor: pipelining
[ a, b, c,...]
[ (echo_1 a), (echo_1 b), (echo_1 c)]
(echo_2 (echo_1 a))
(echo_2 (echo_1 b))
(echo_2 (echo_1 c))
But:
no “chunking”
no repeated stateful calls to processors
no ranking
14](https://image.slidesharecdn.com/seco-workshop-paolo-100622043916-phpapp02/75/Invited-talk-Second-Search-Computing-workshop-28-2048.jpg)