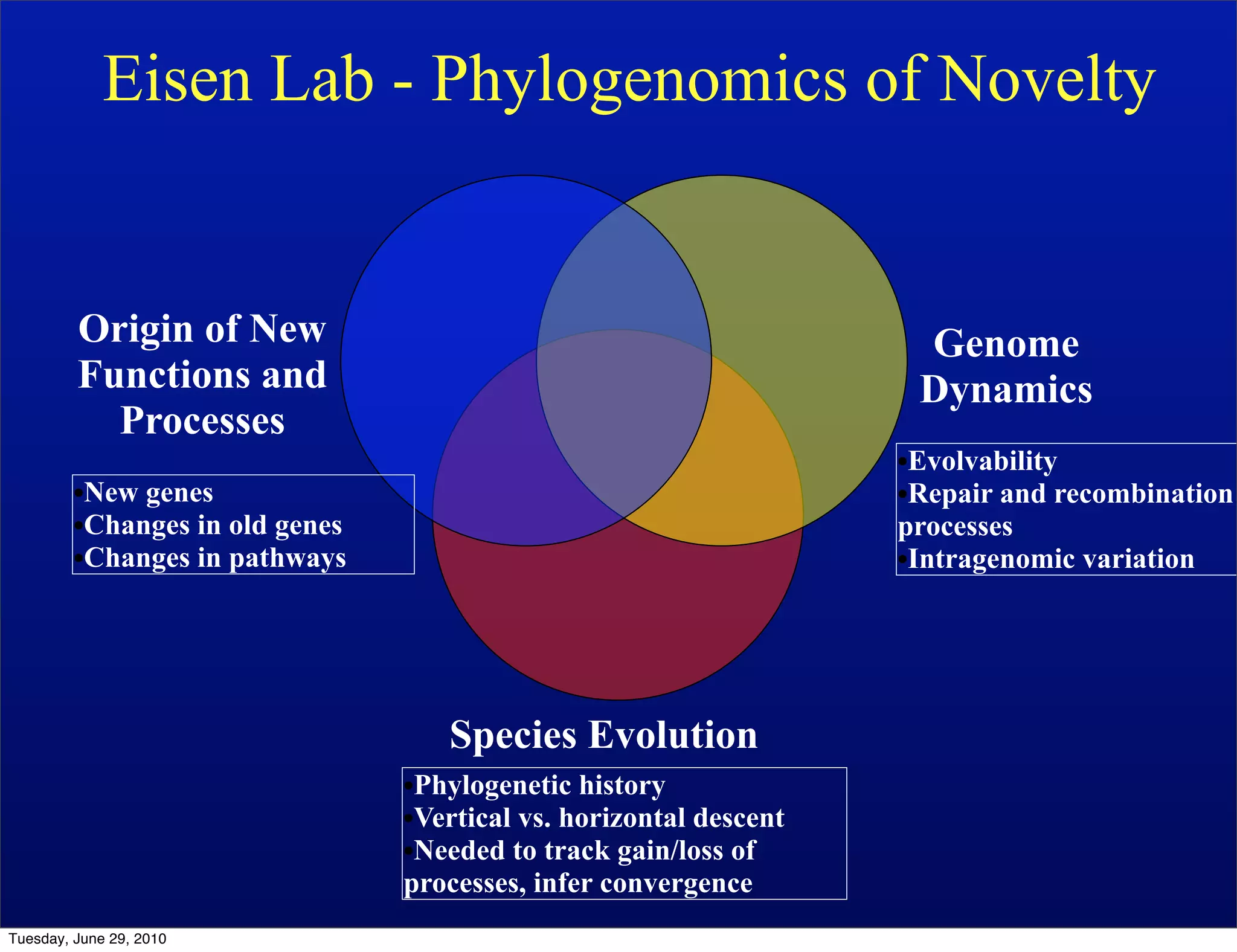
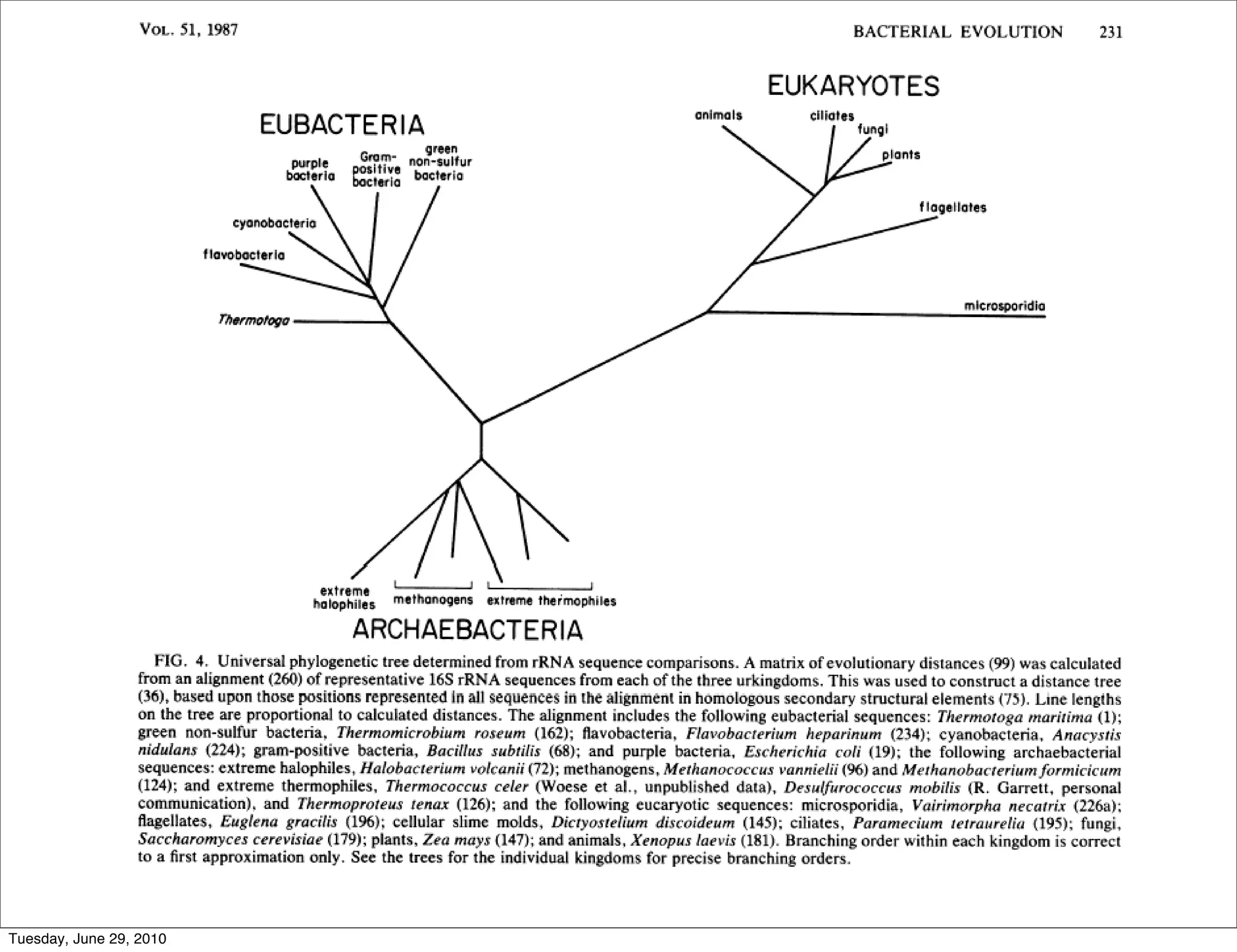
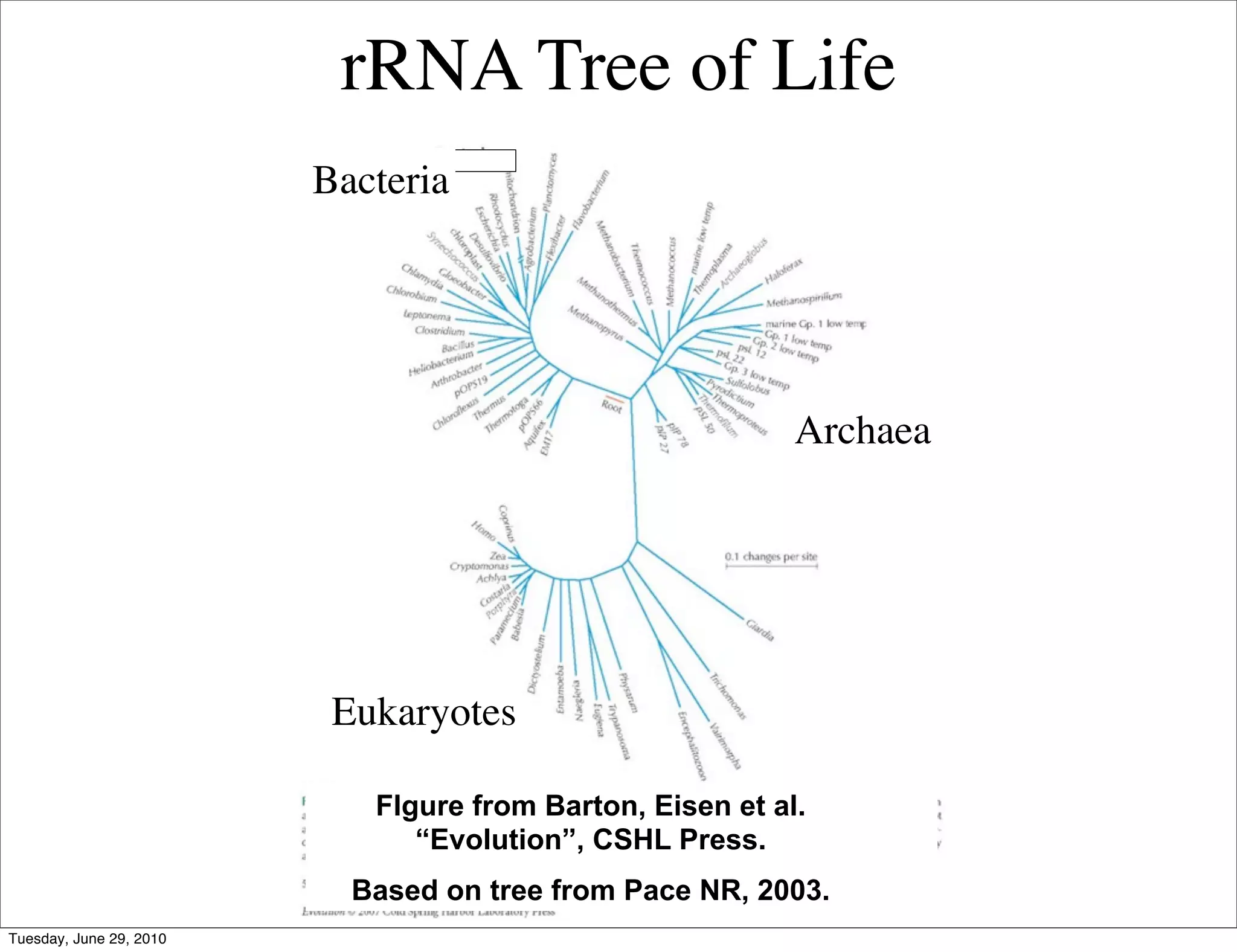
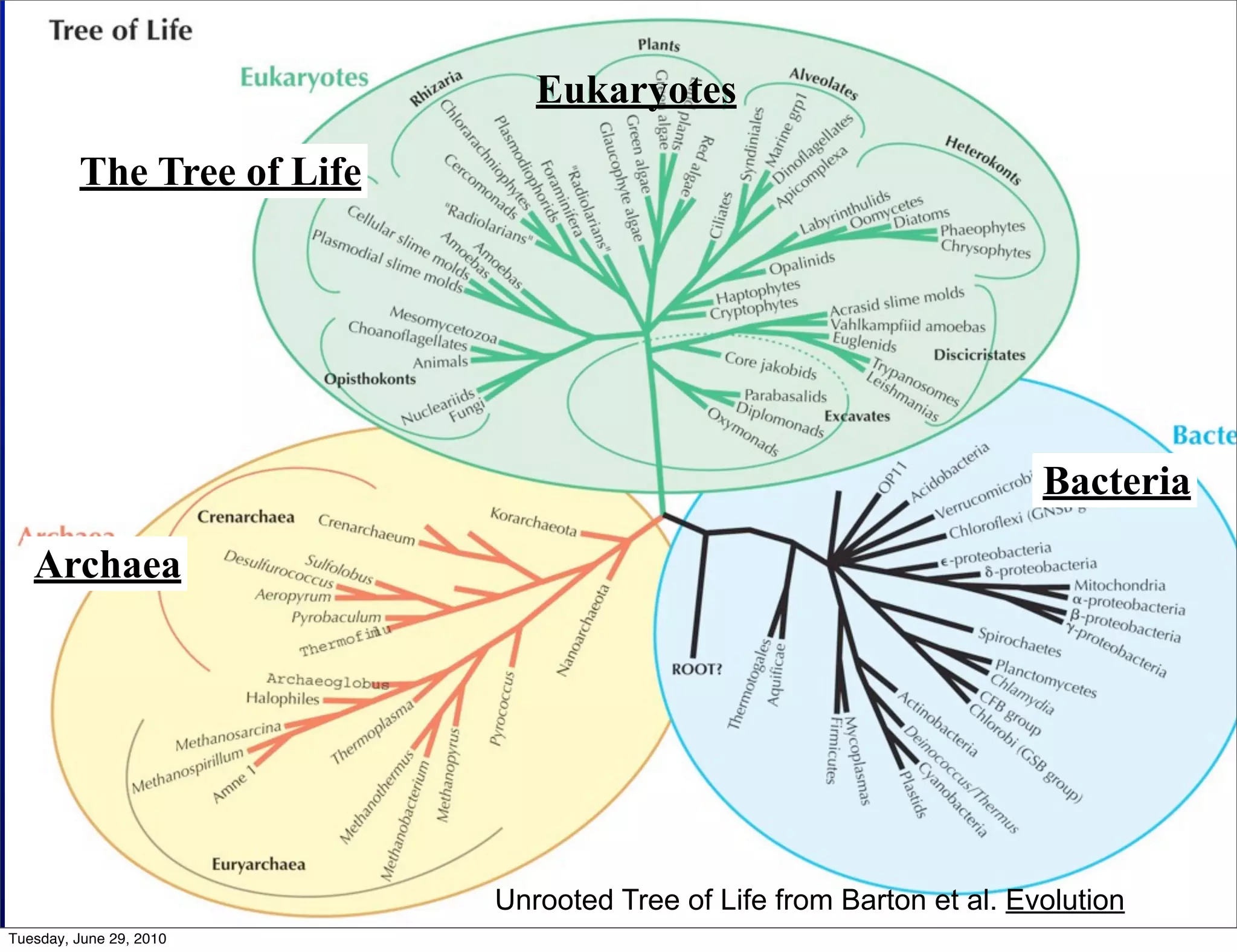
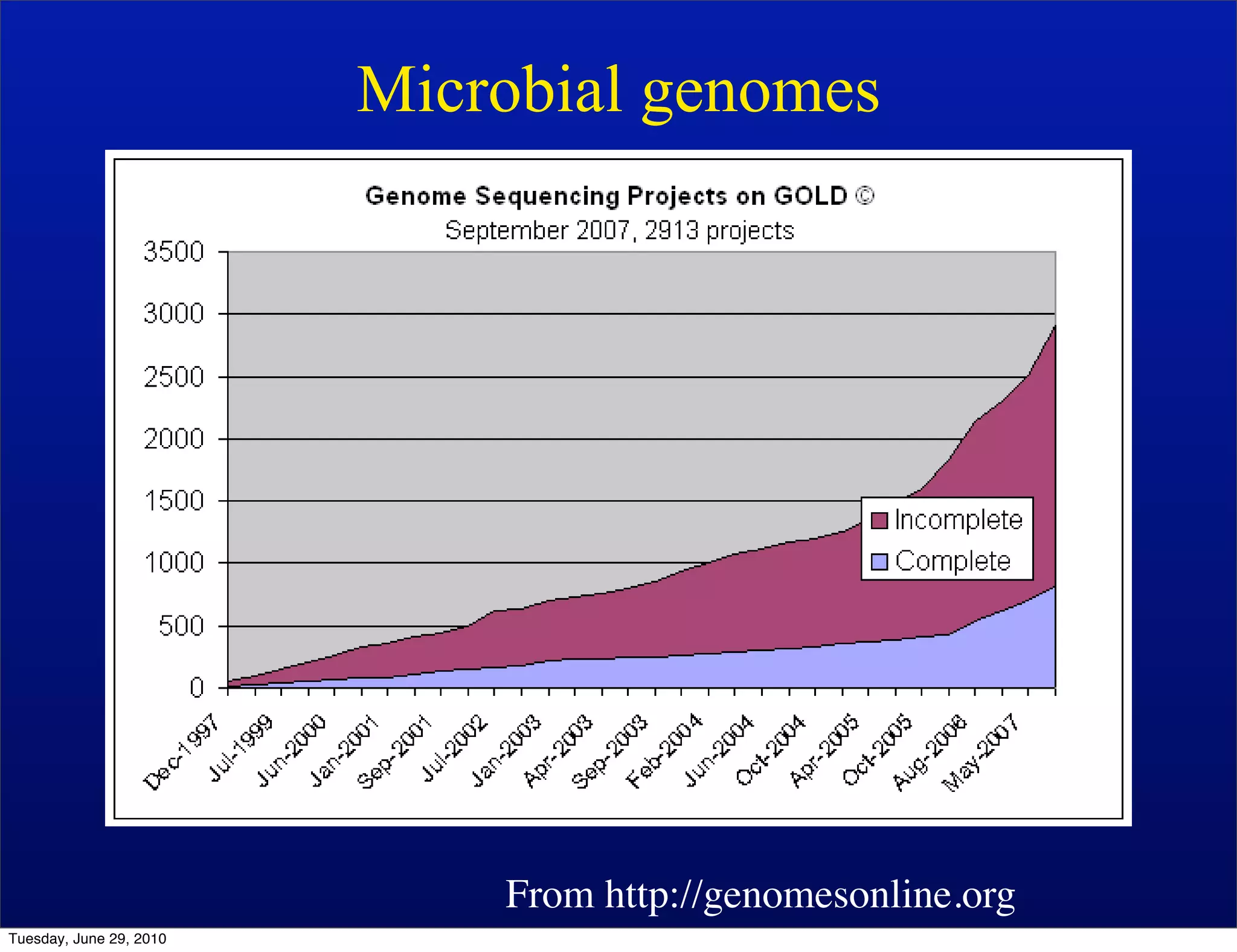
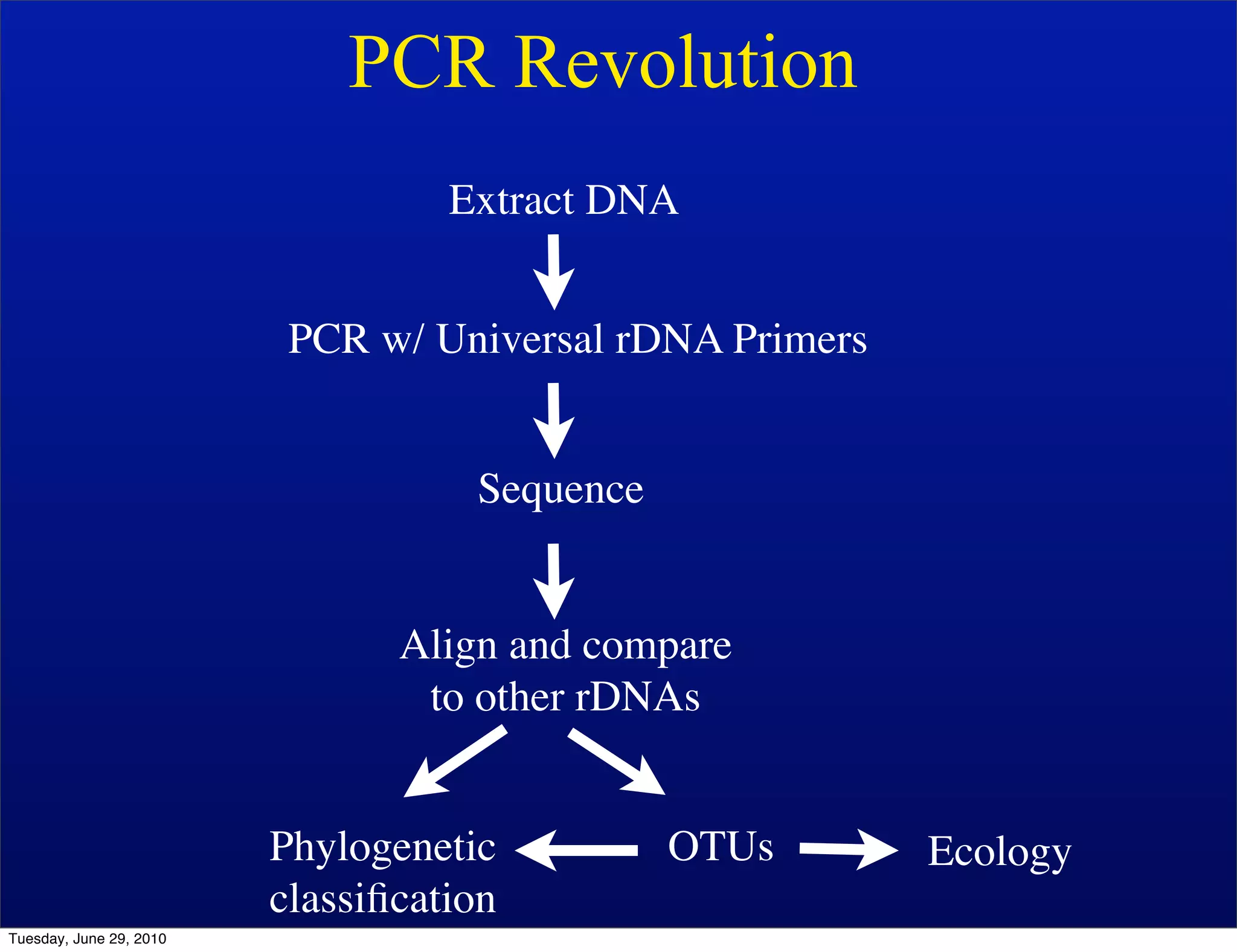
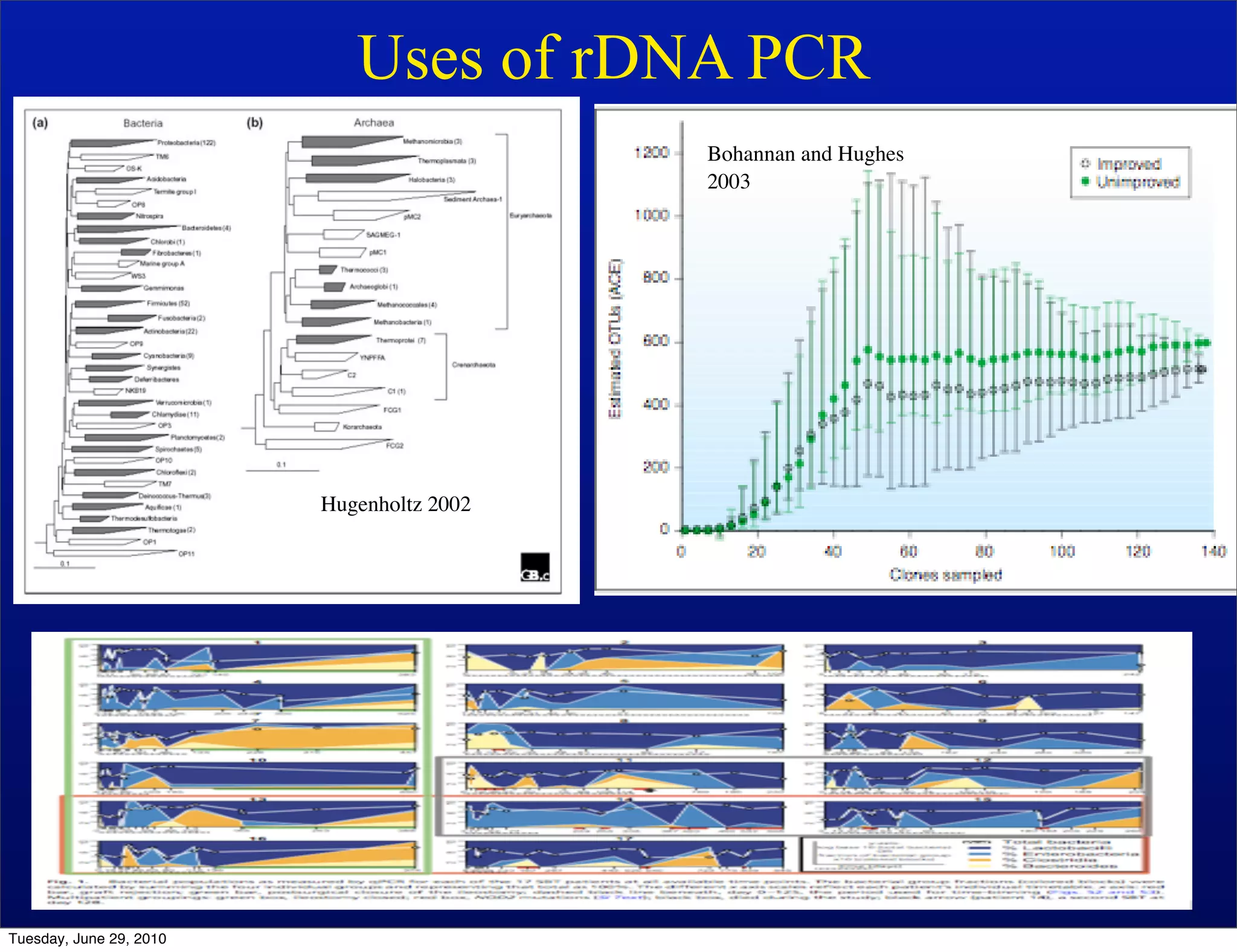
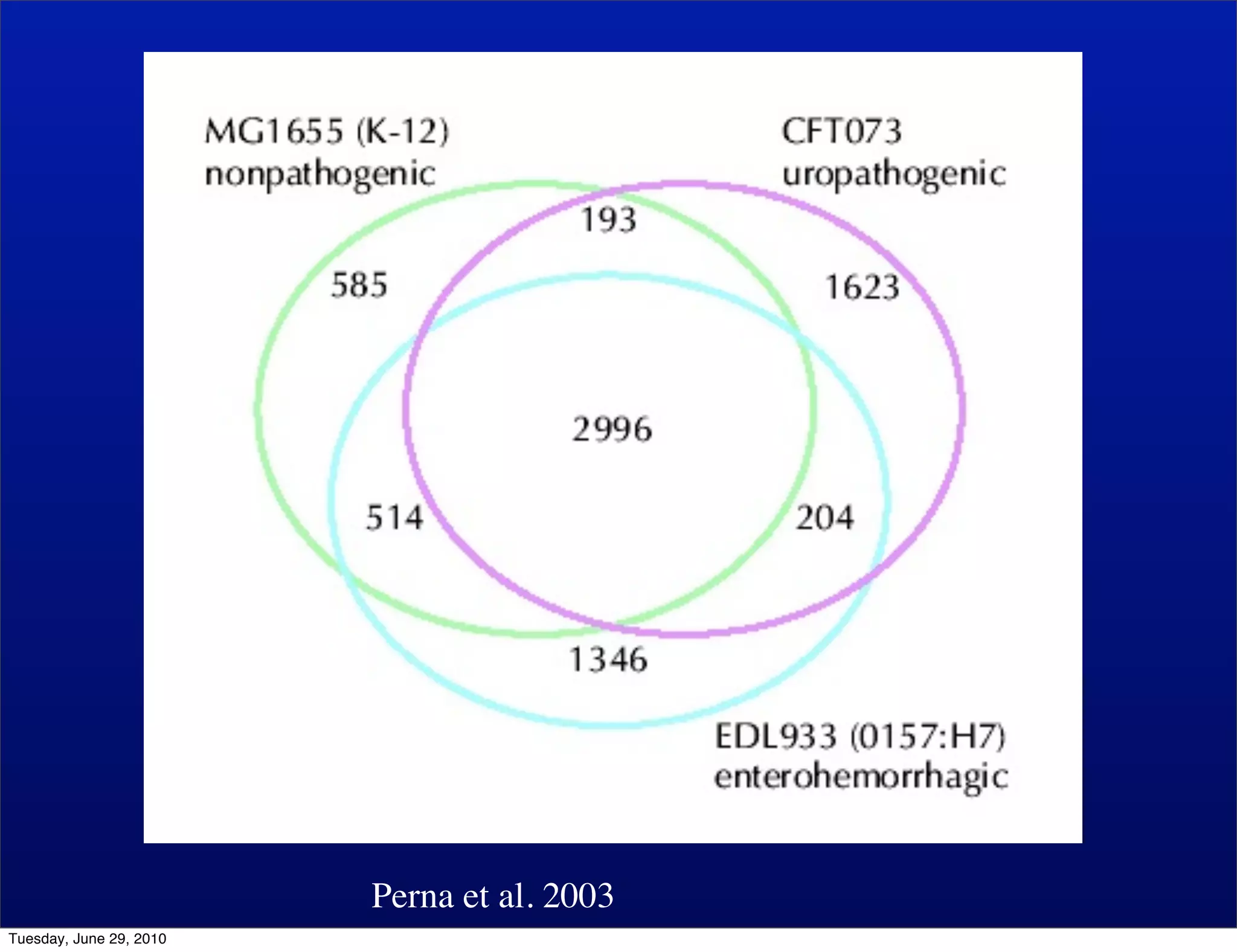
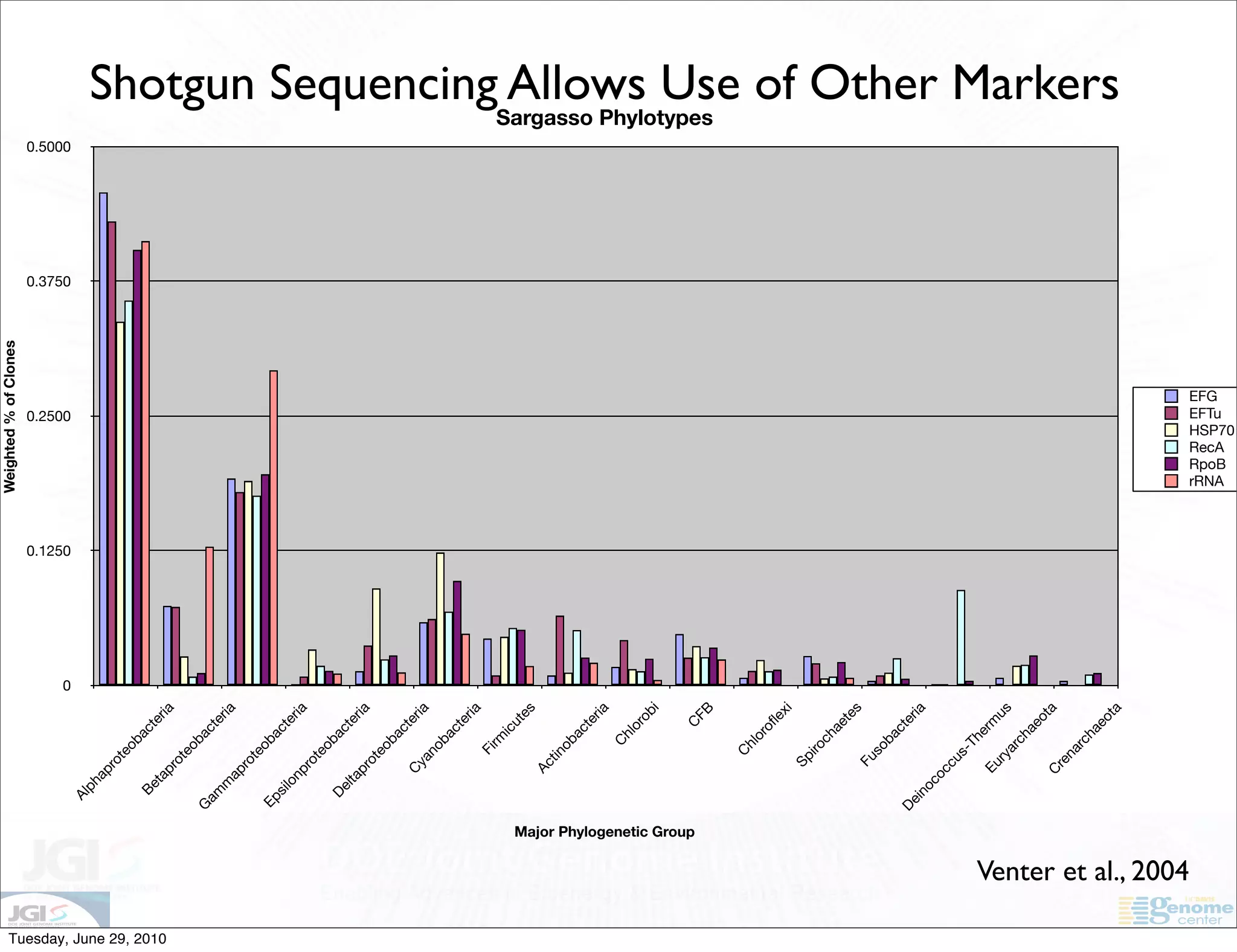
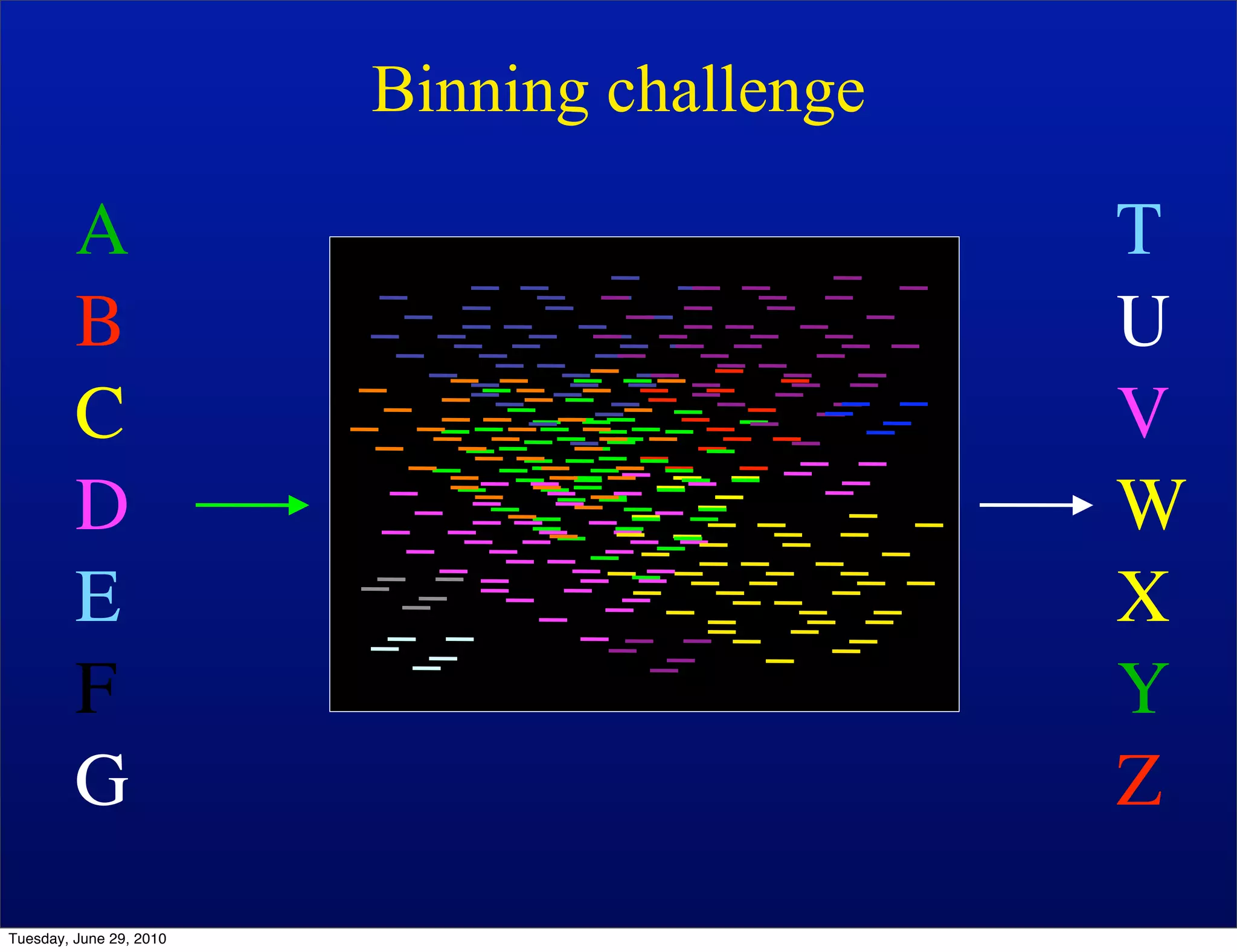
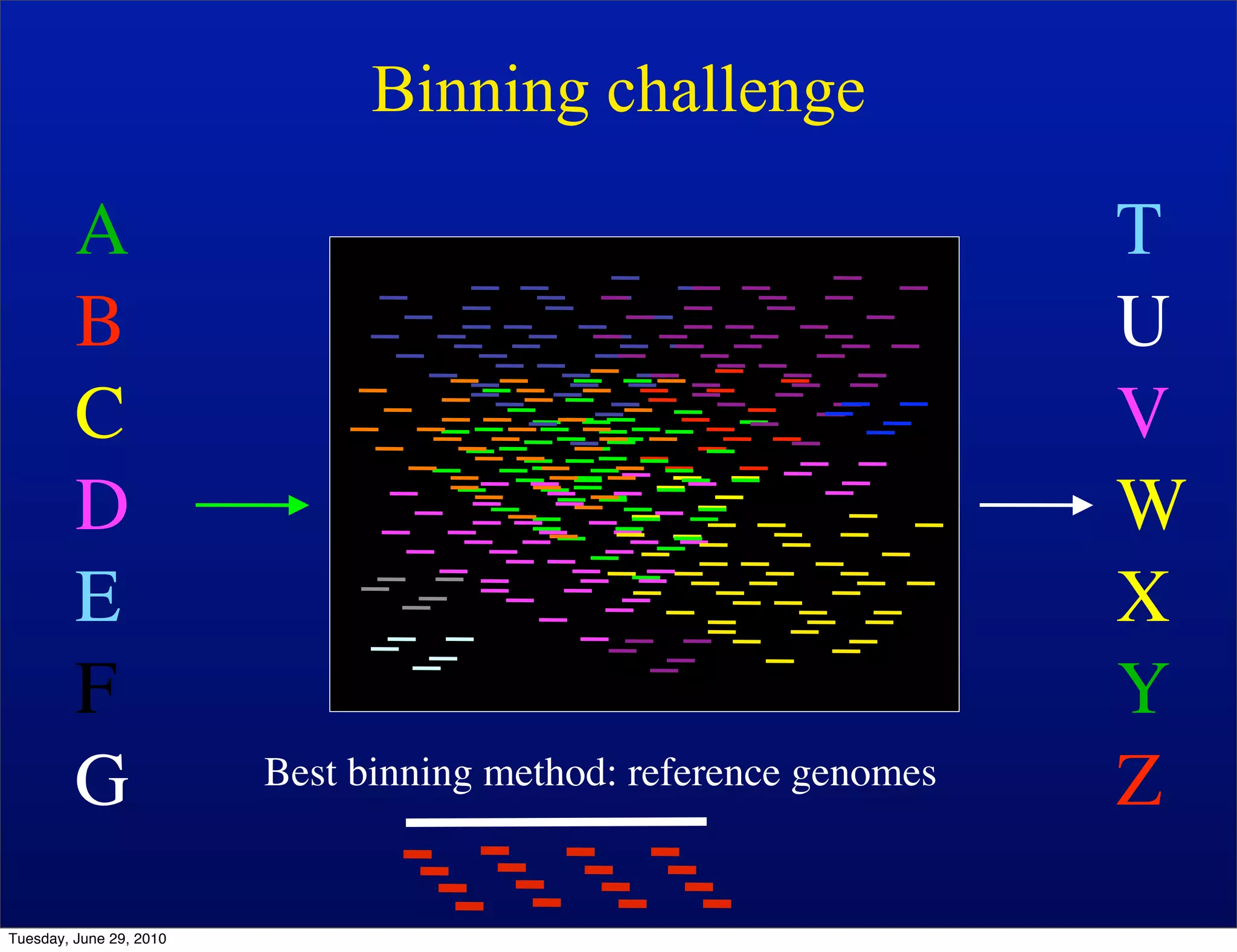
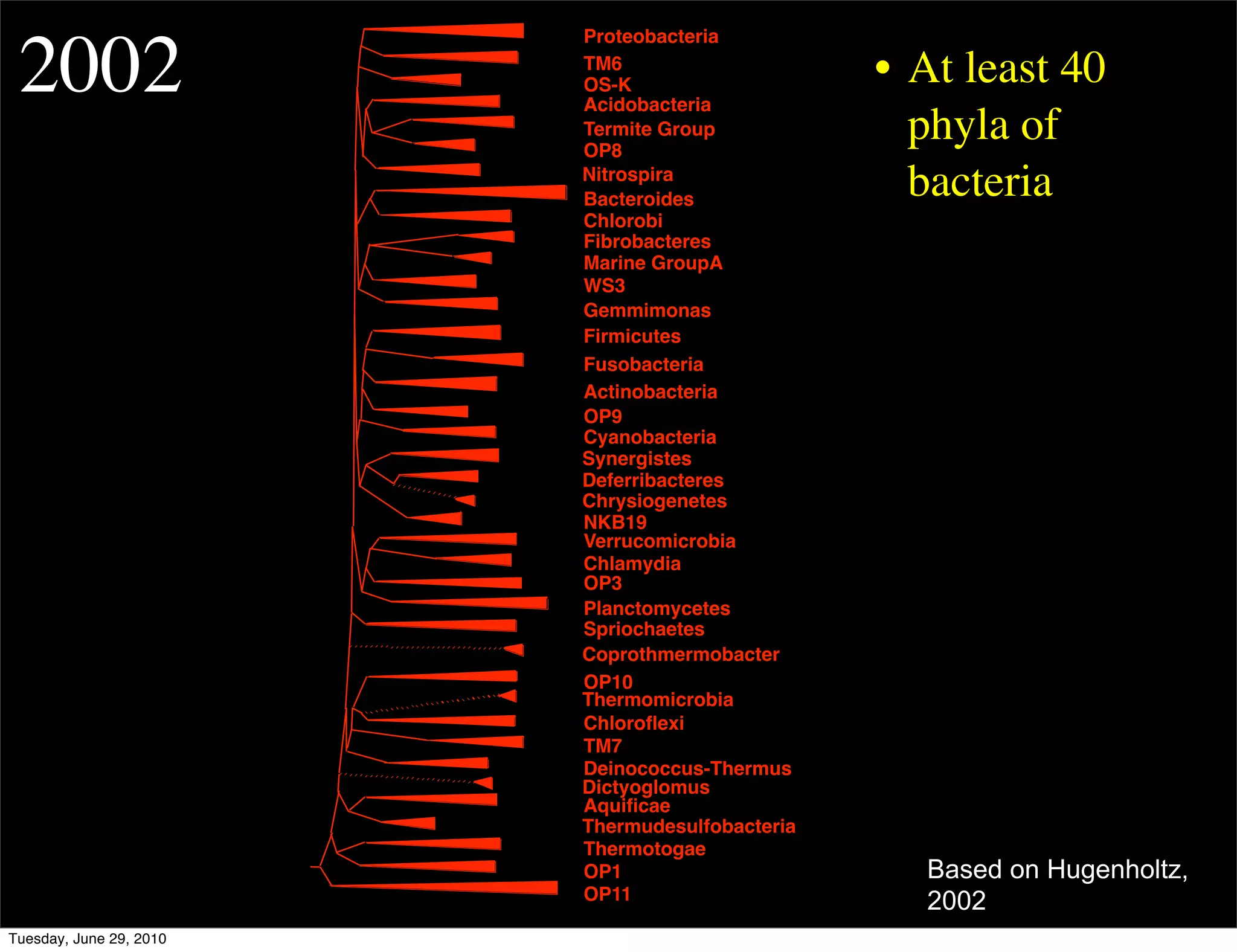
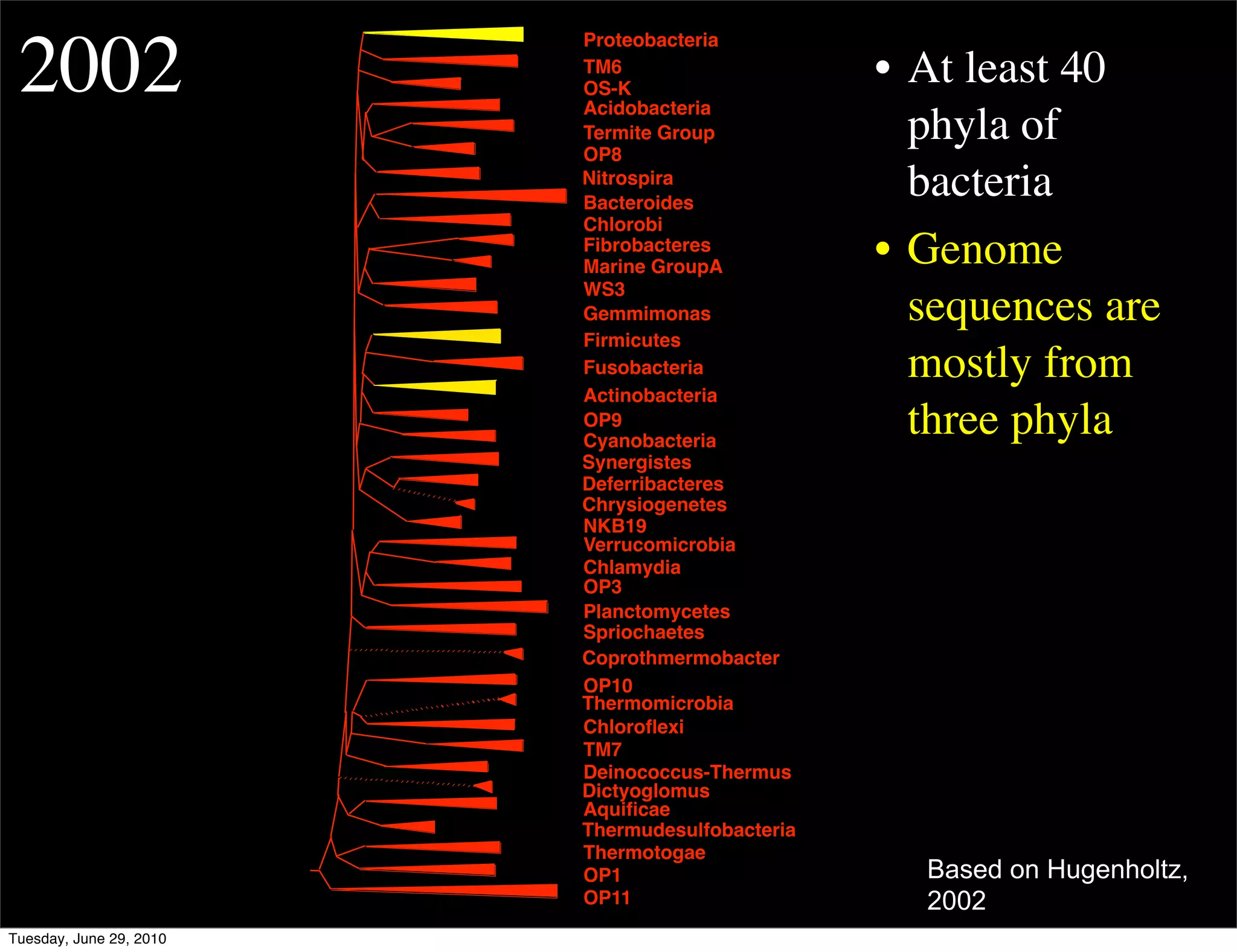
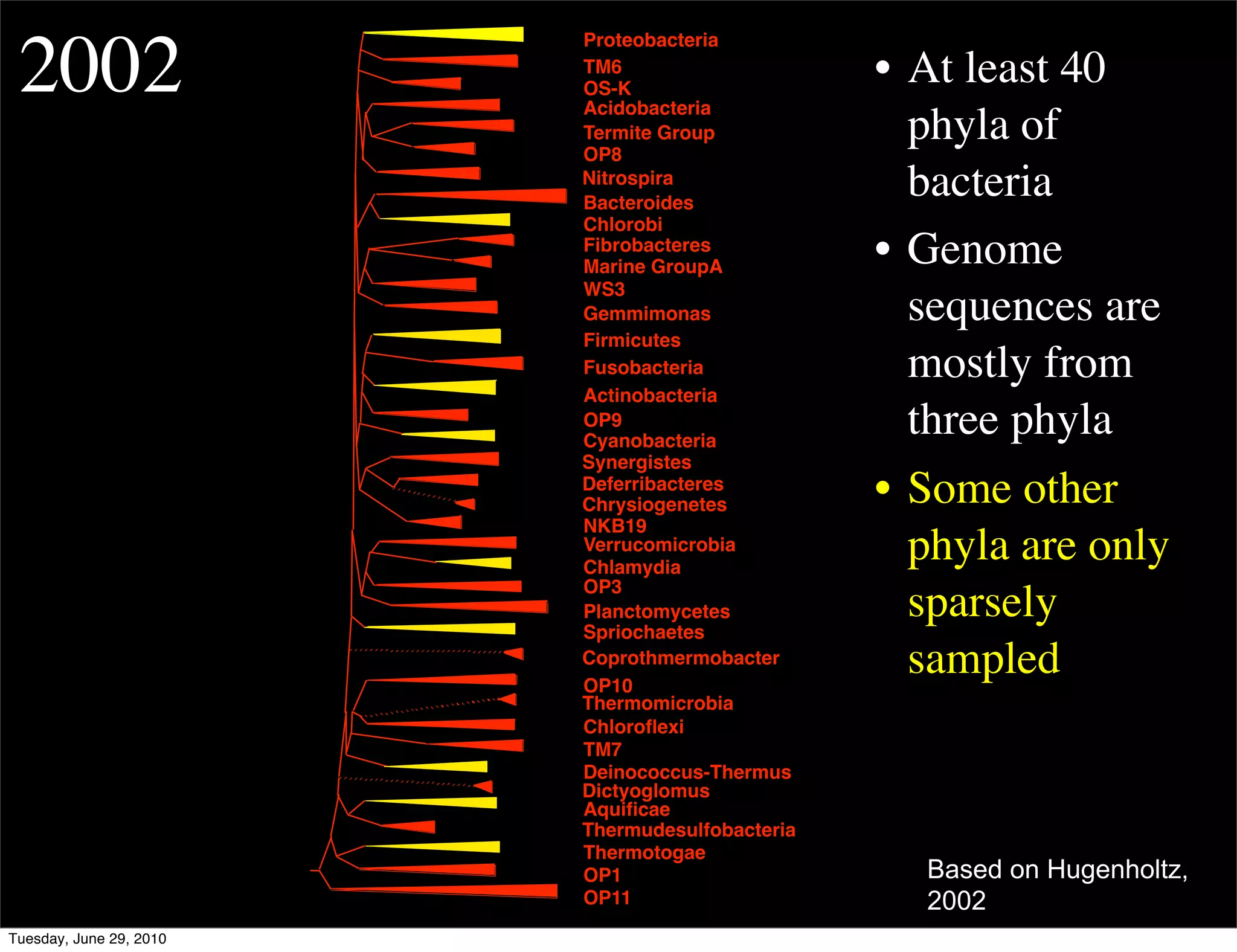
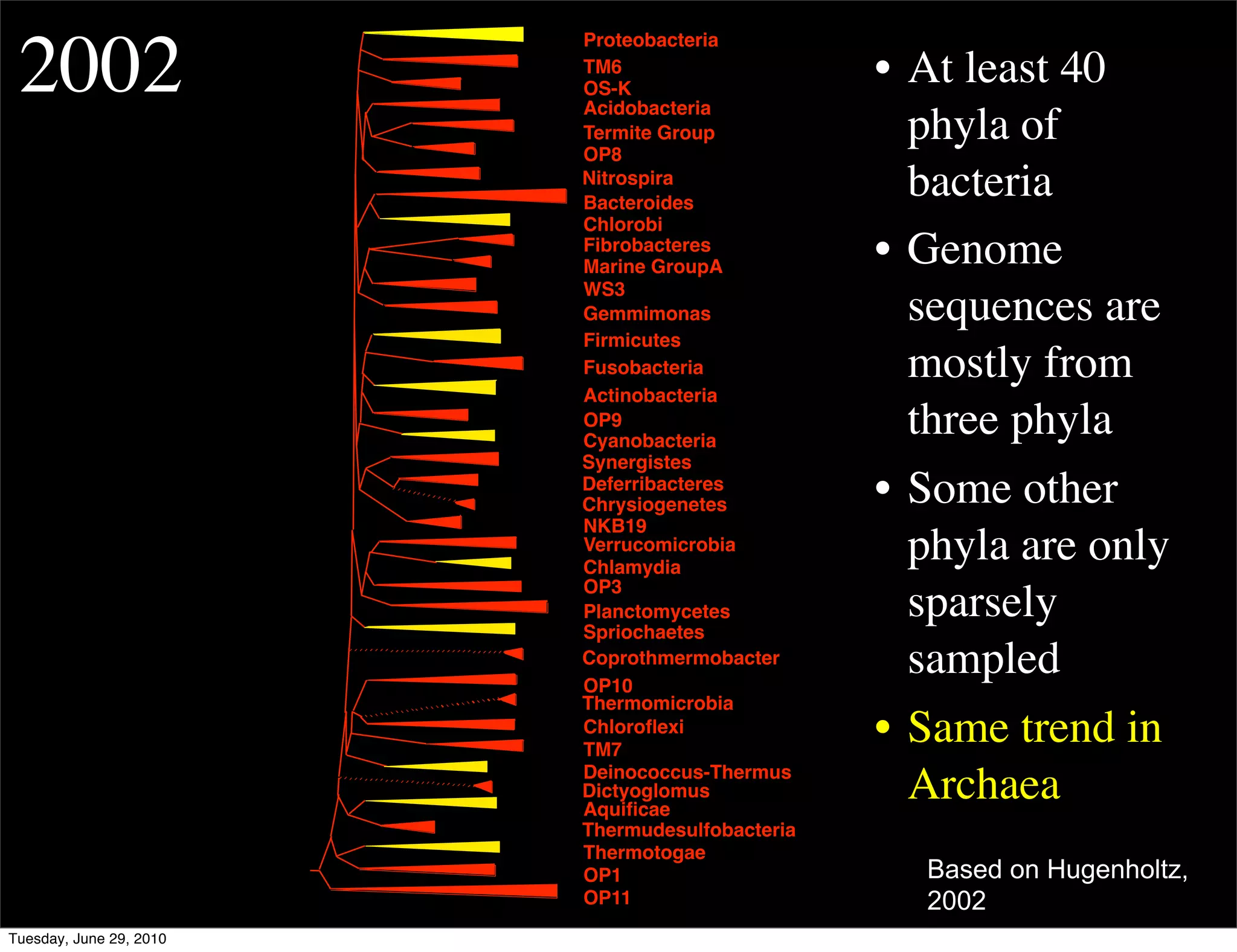
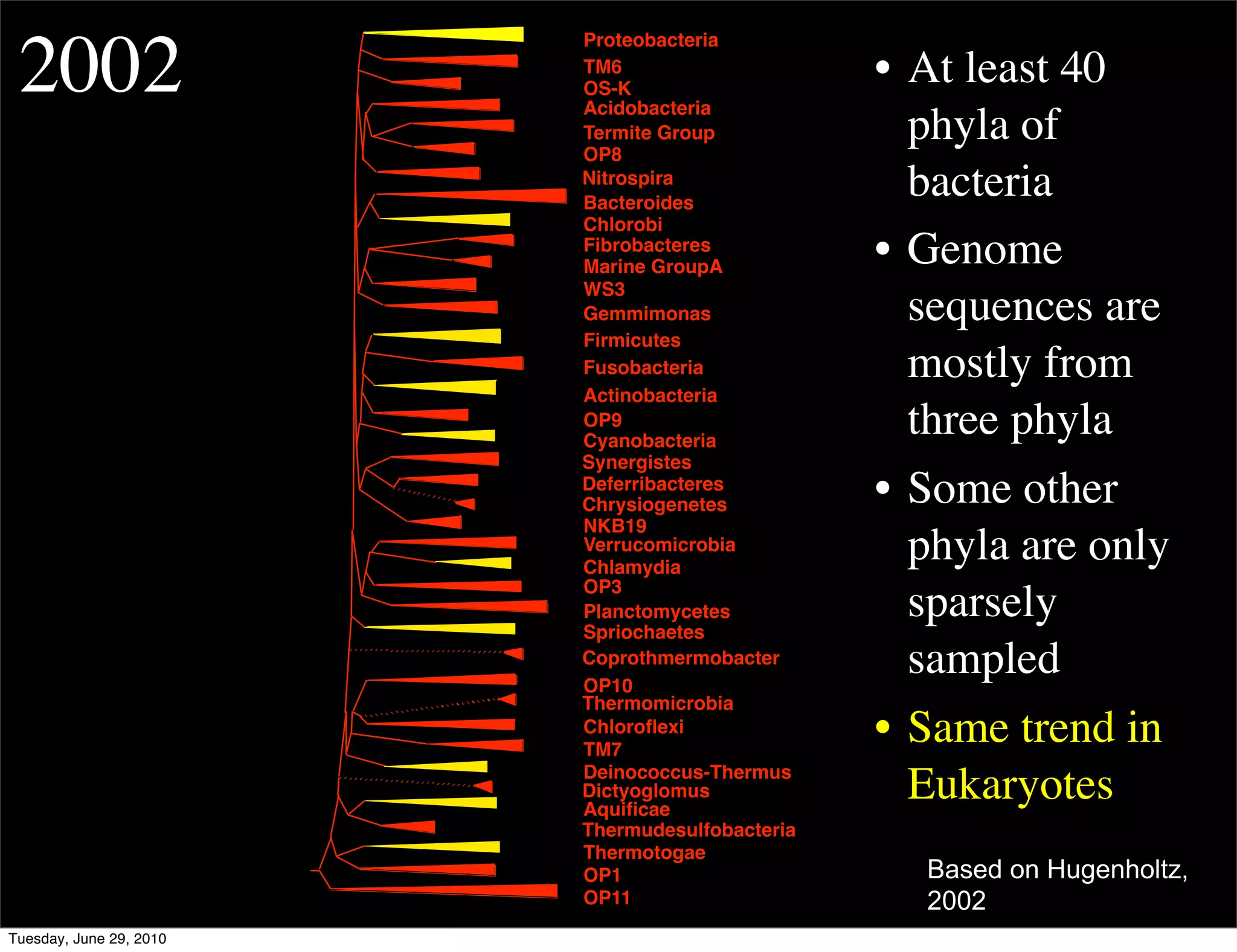
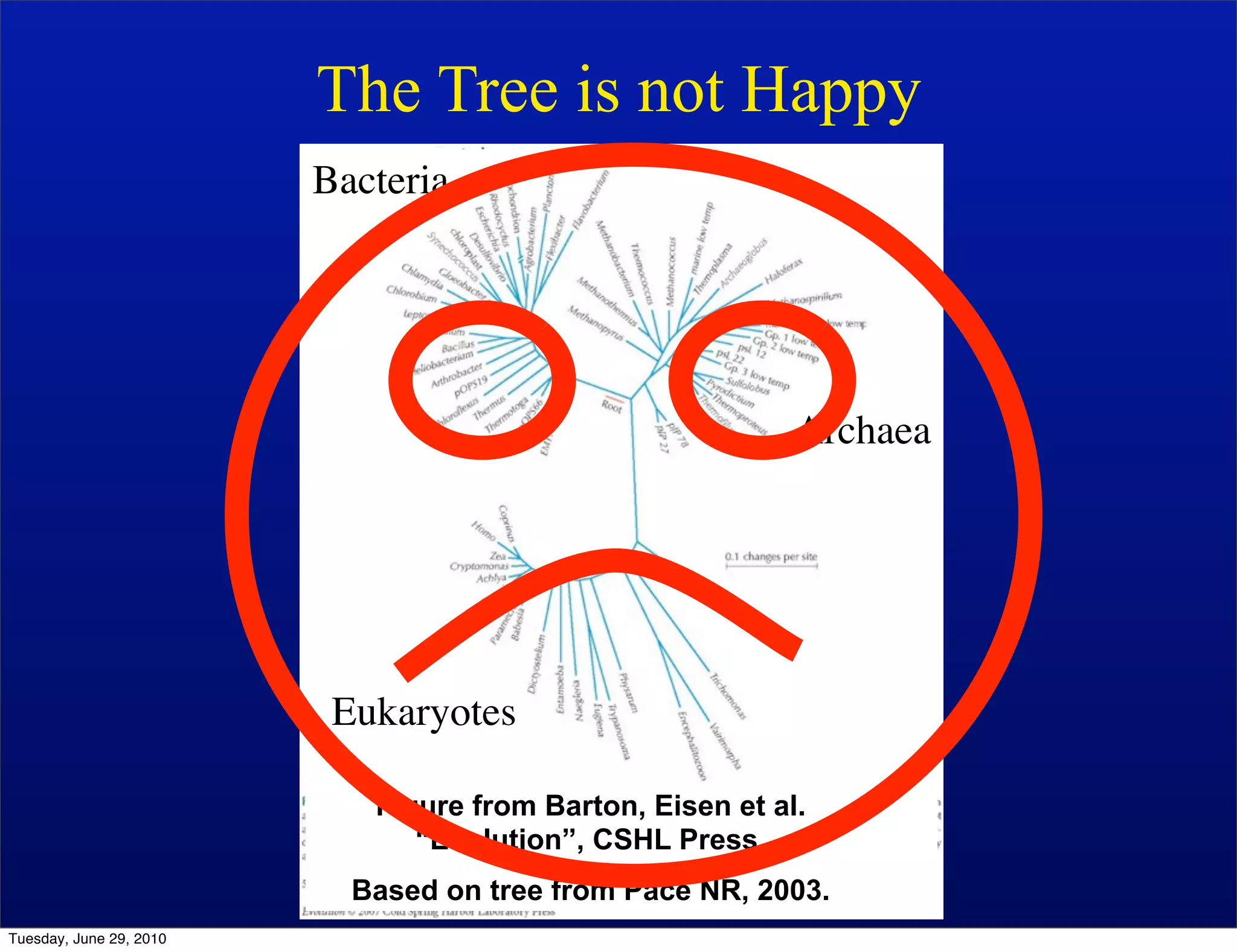
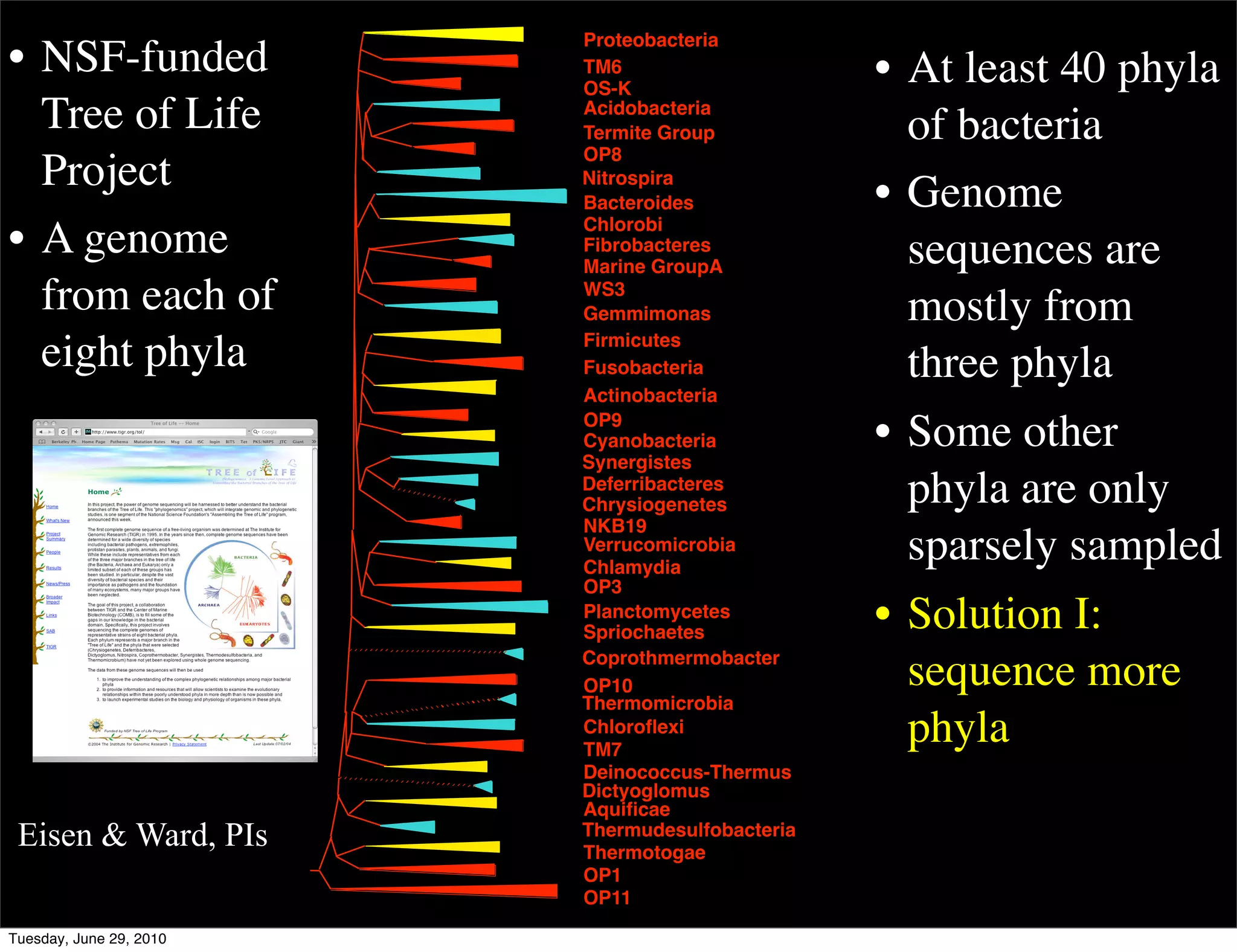
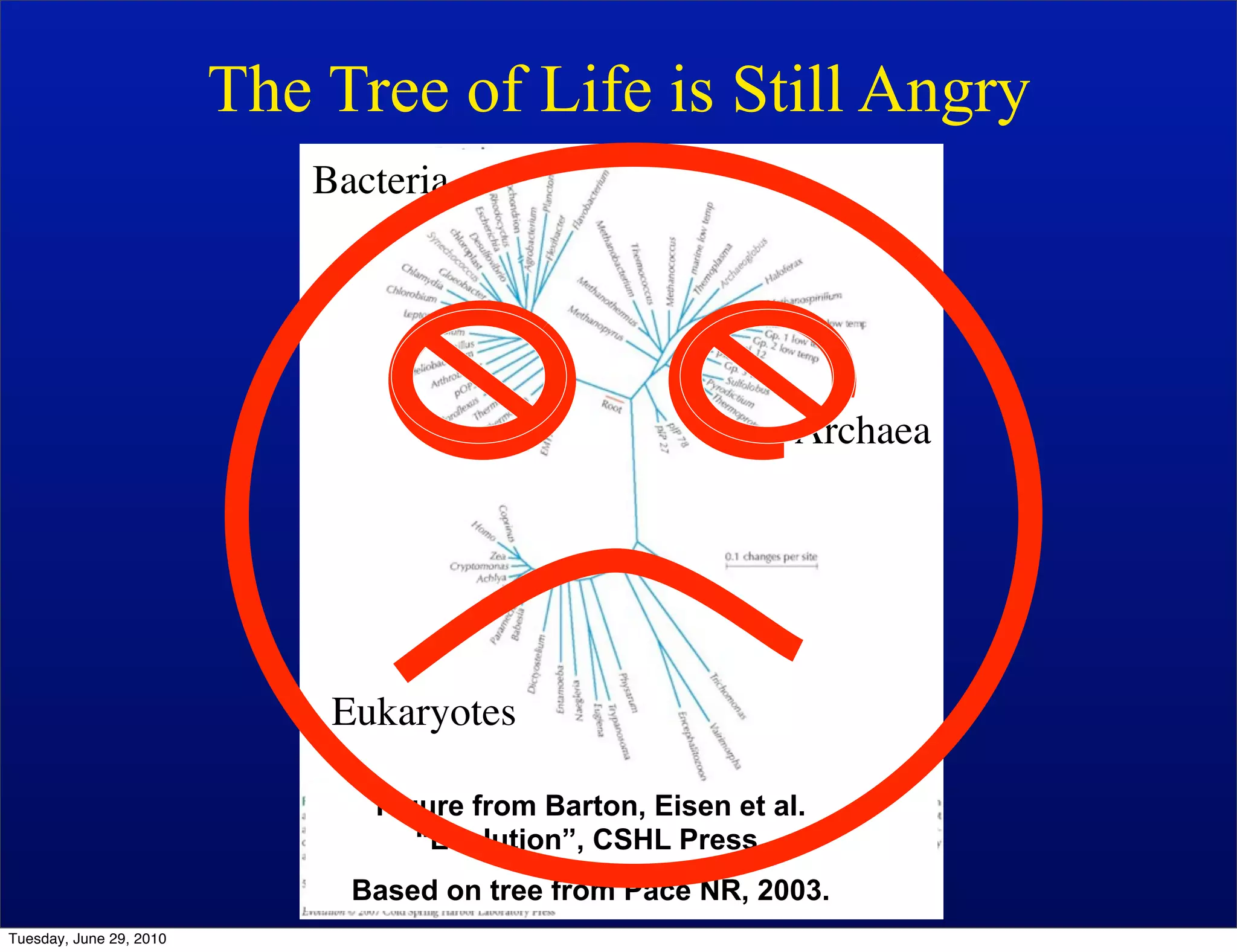
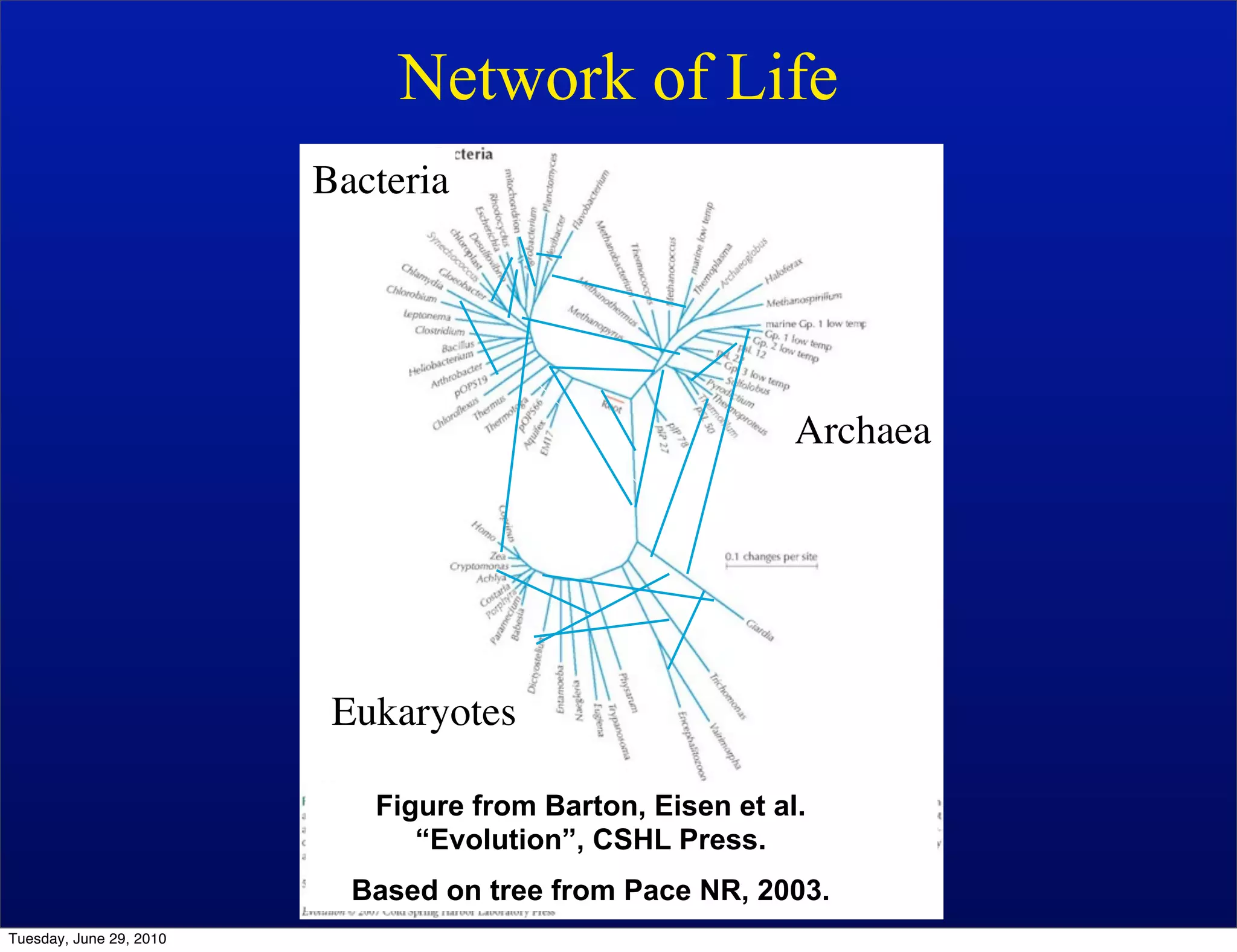
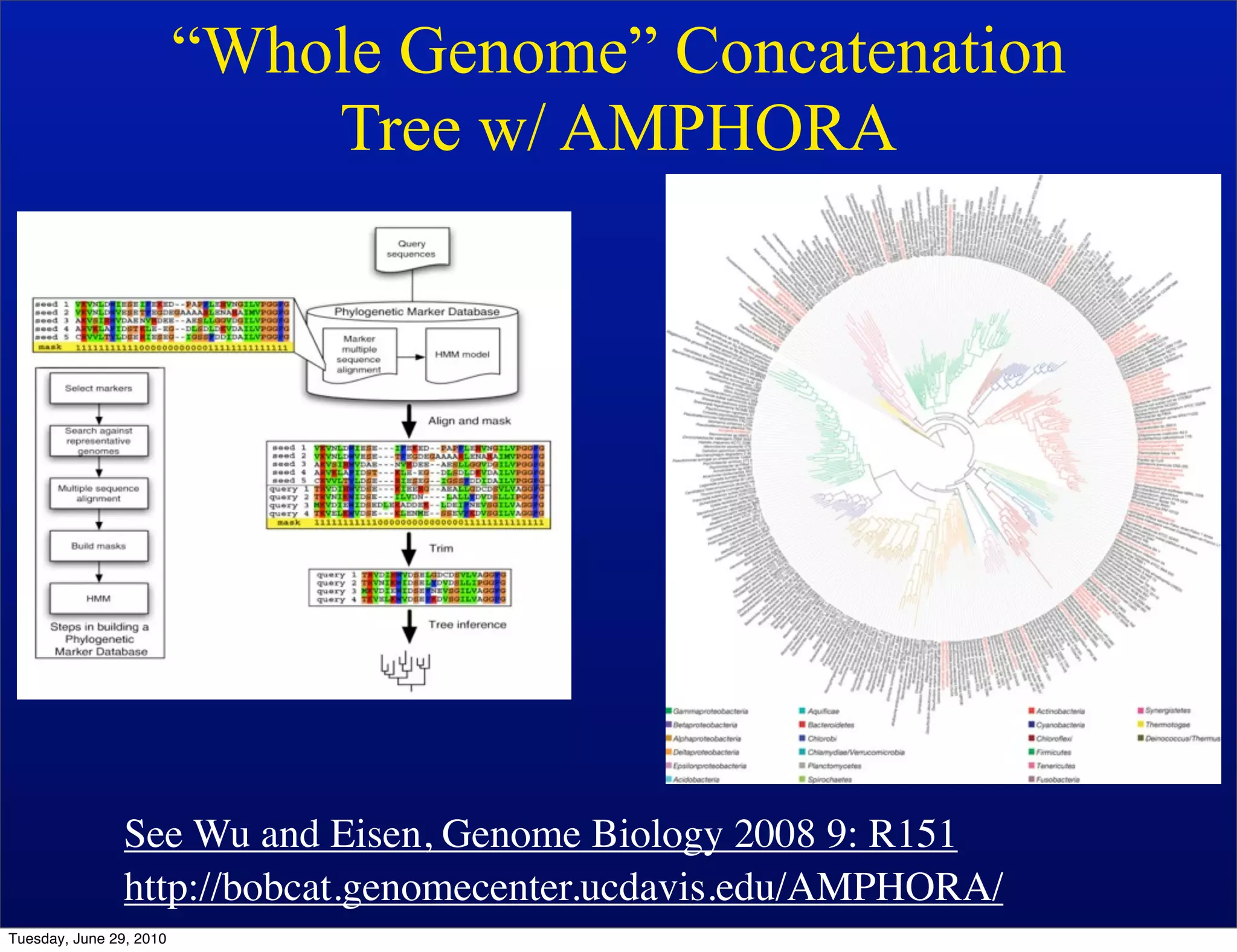
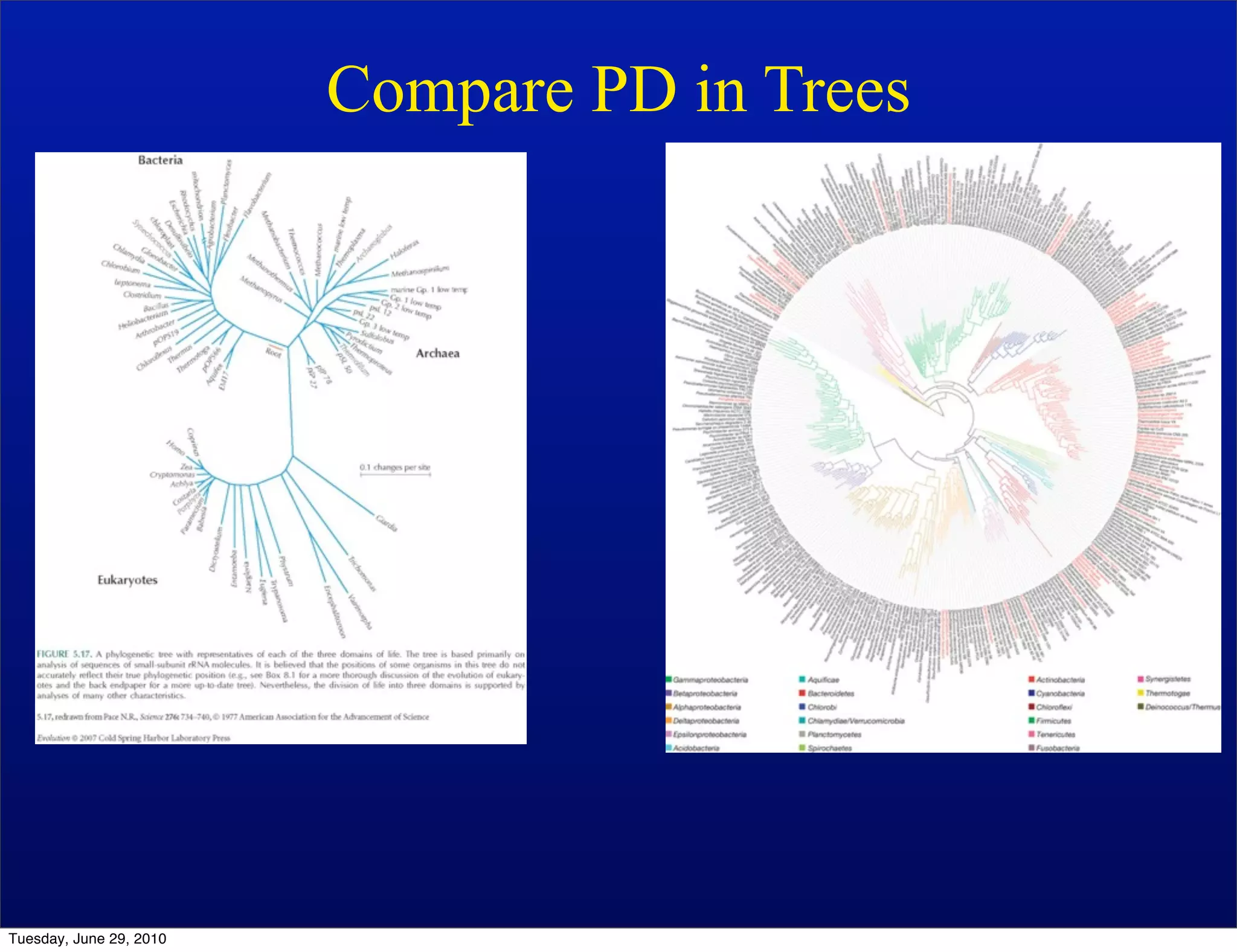
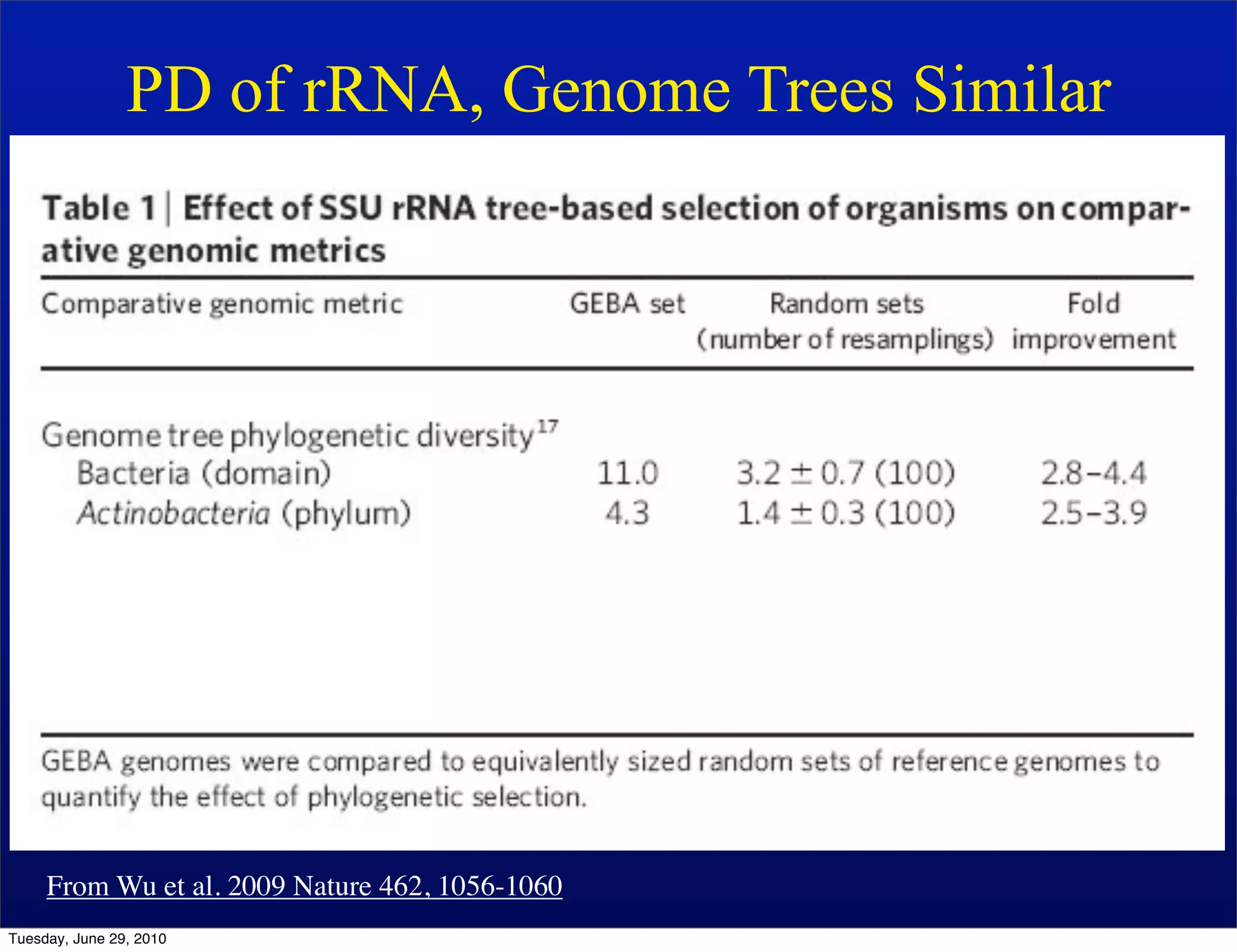
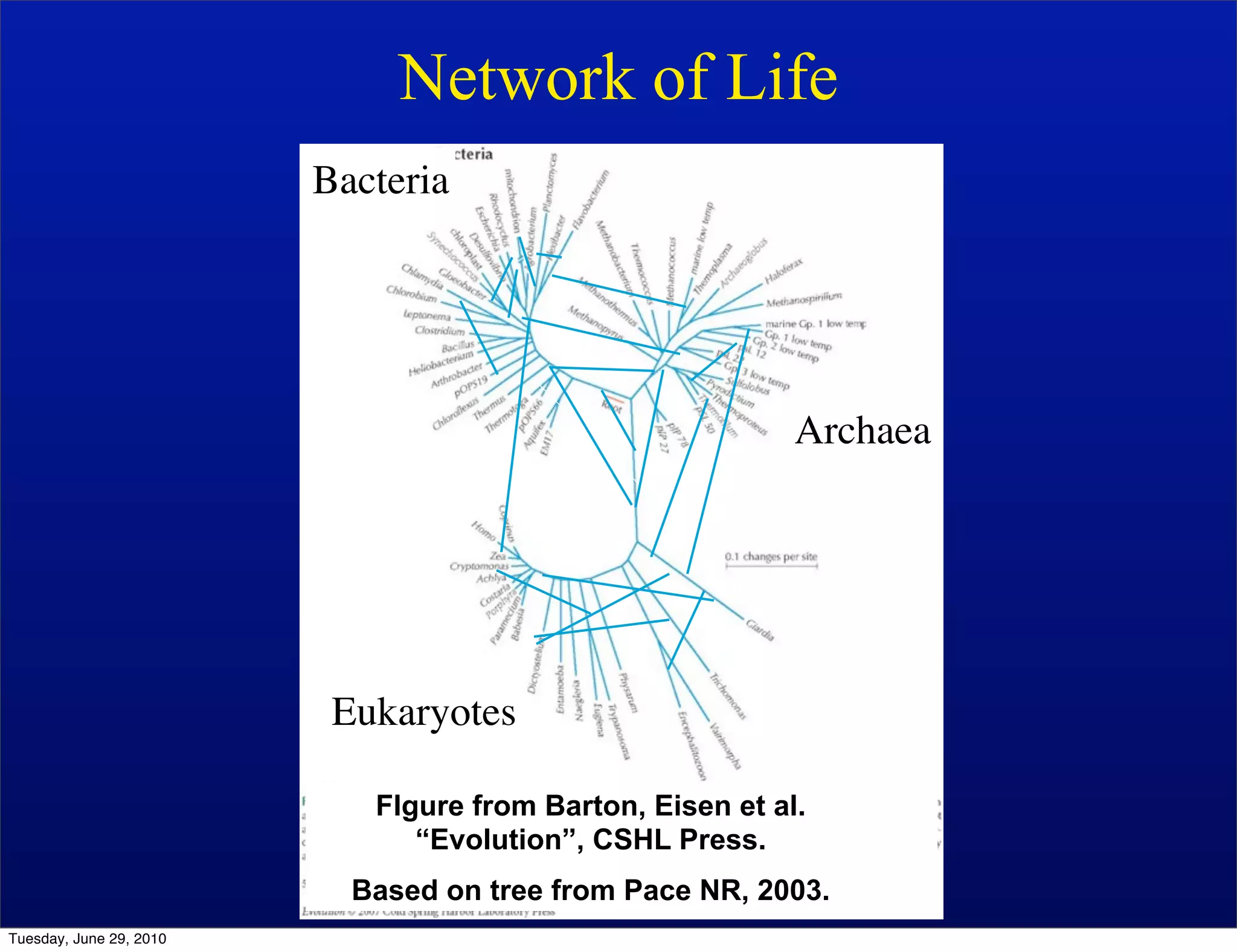
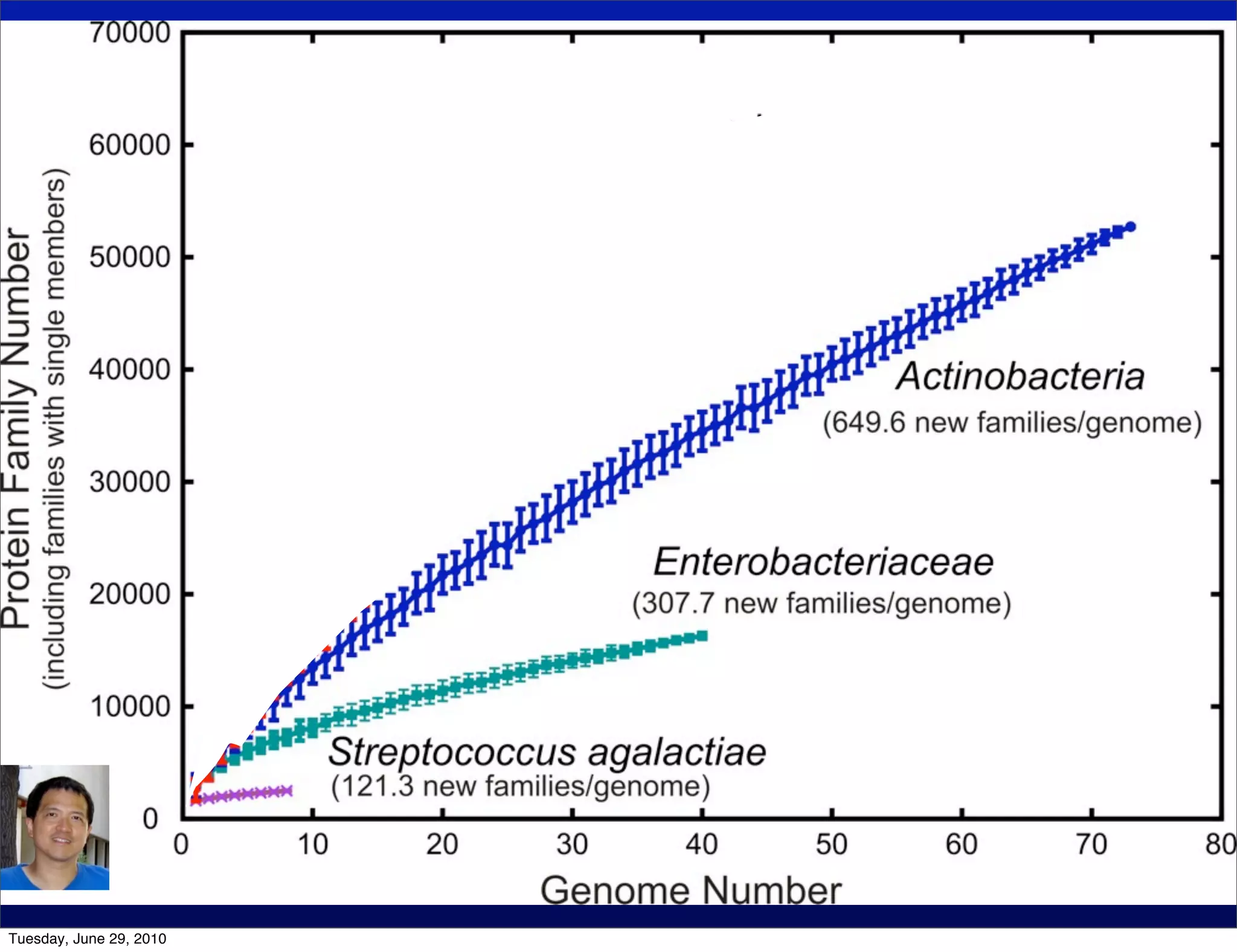
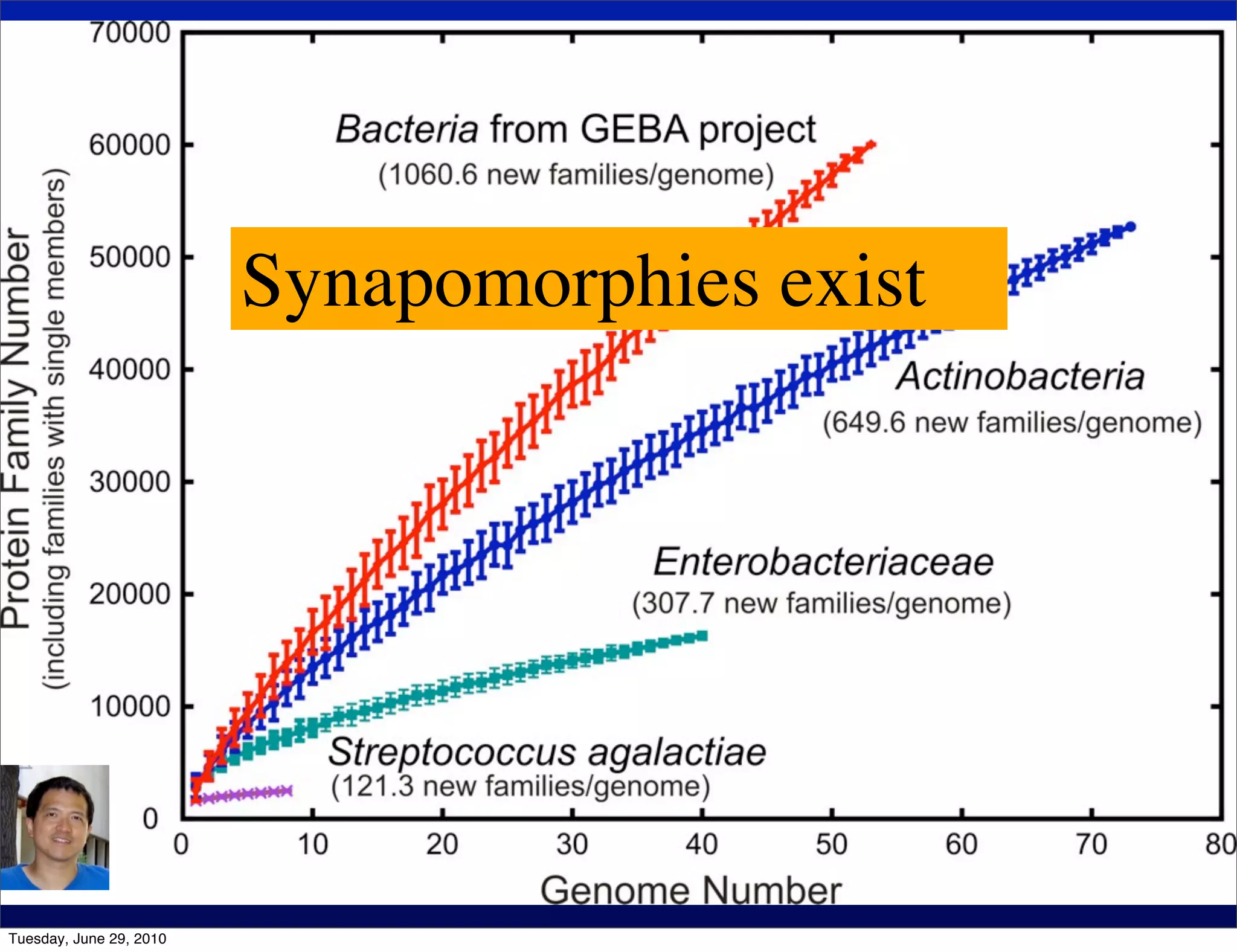
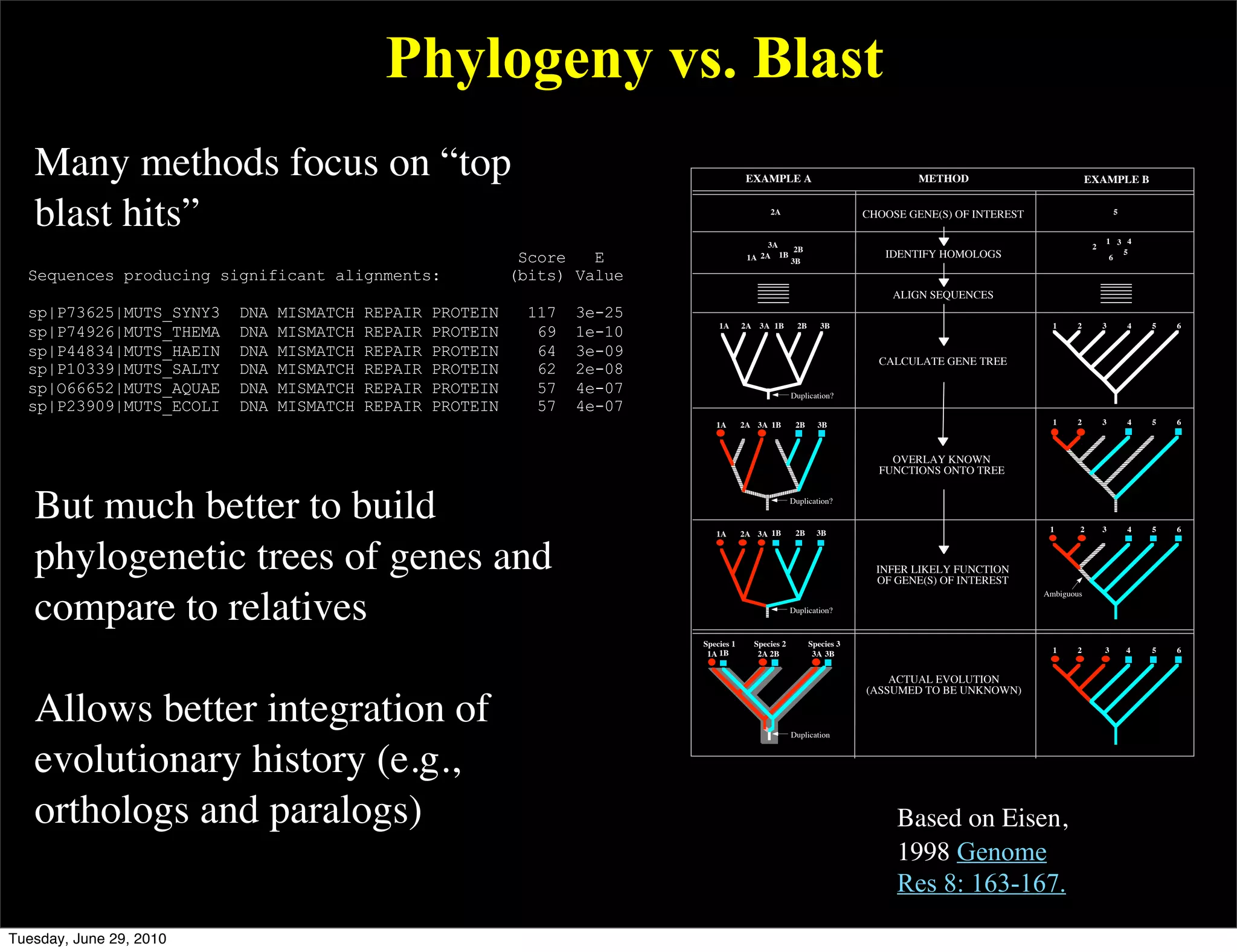
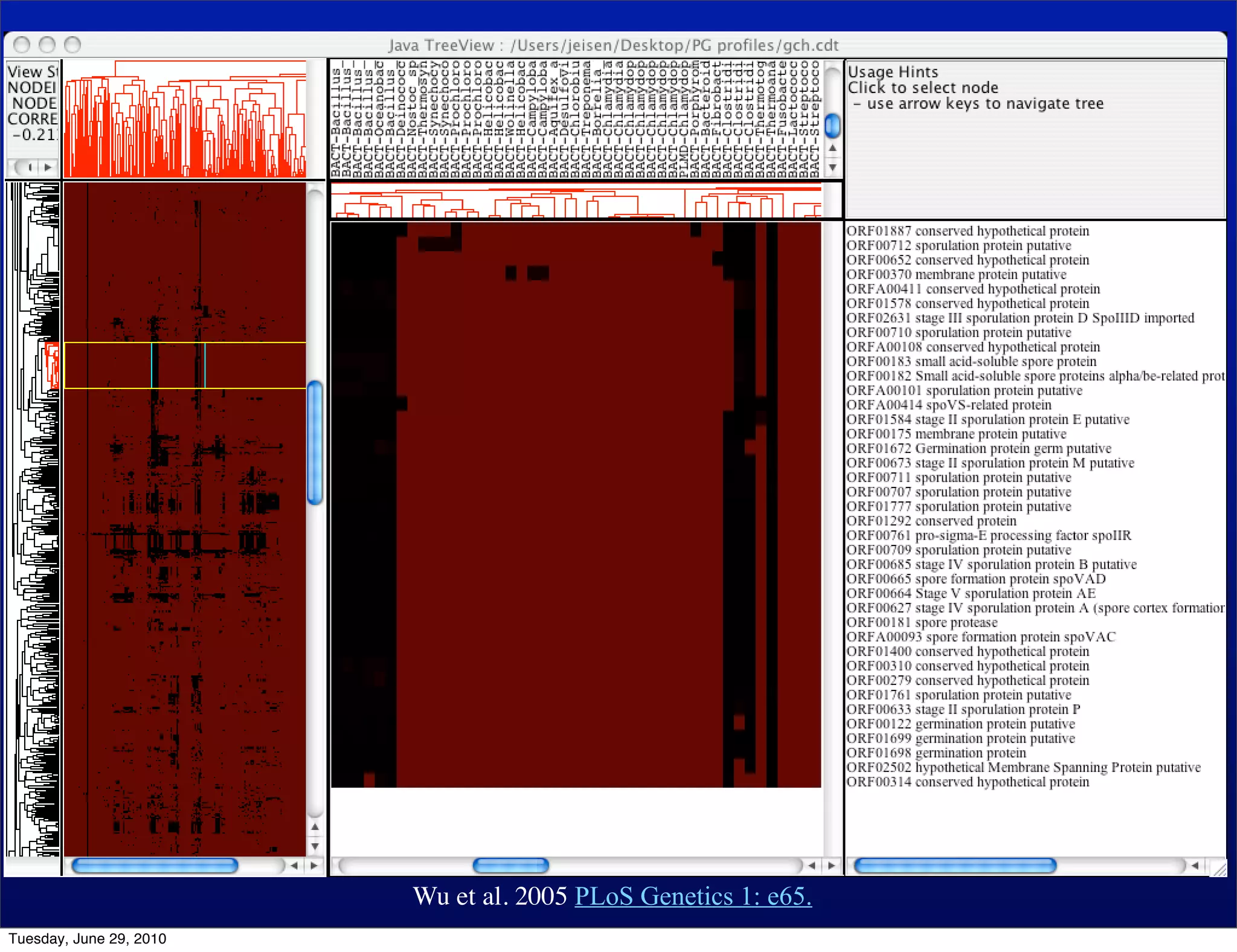
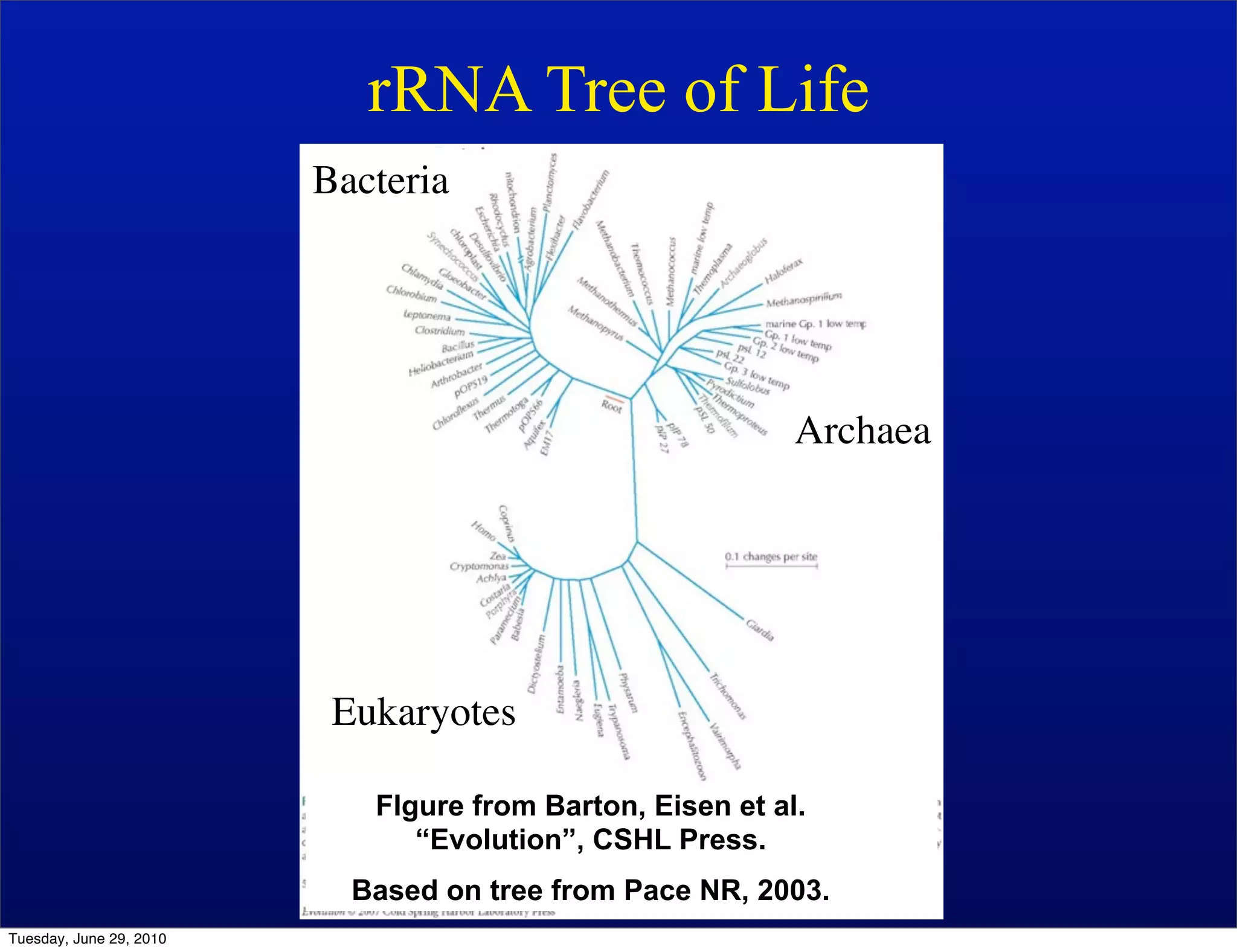
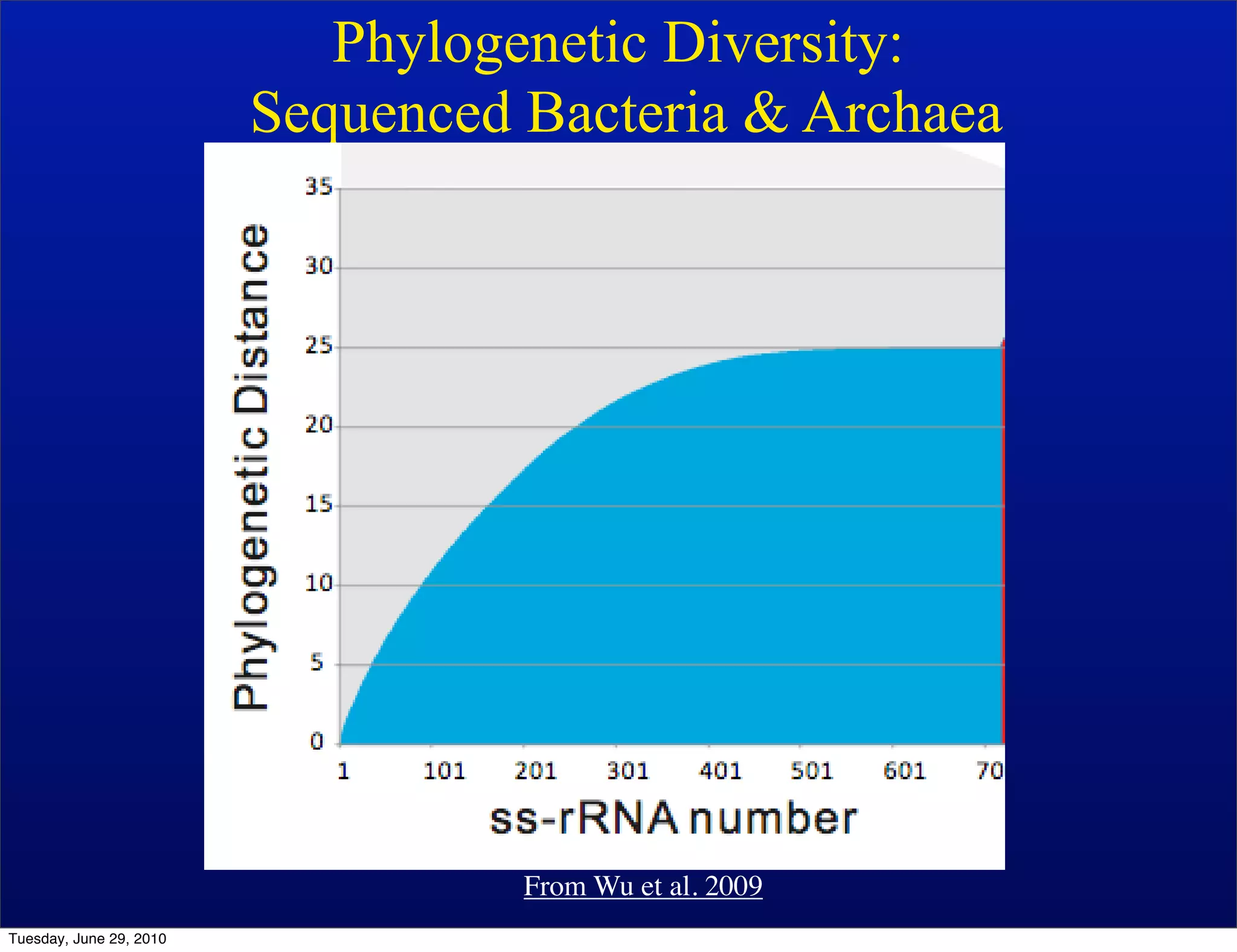
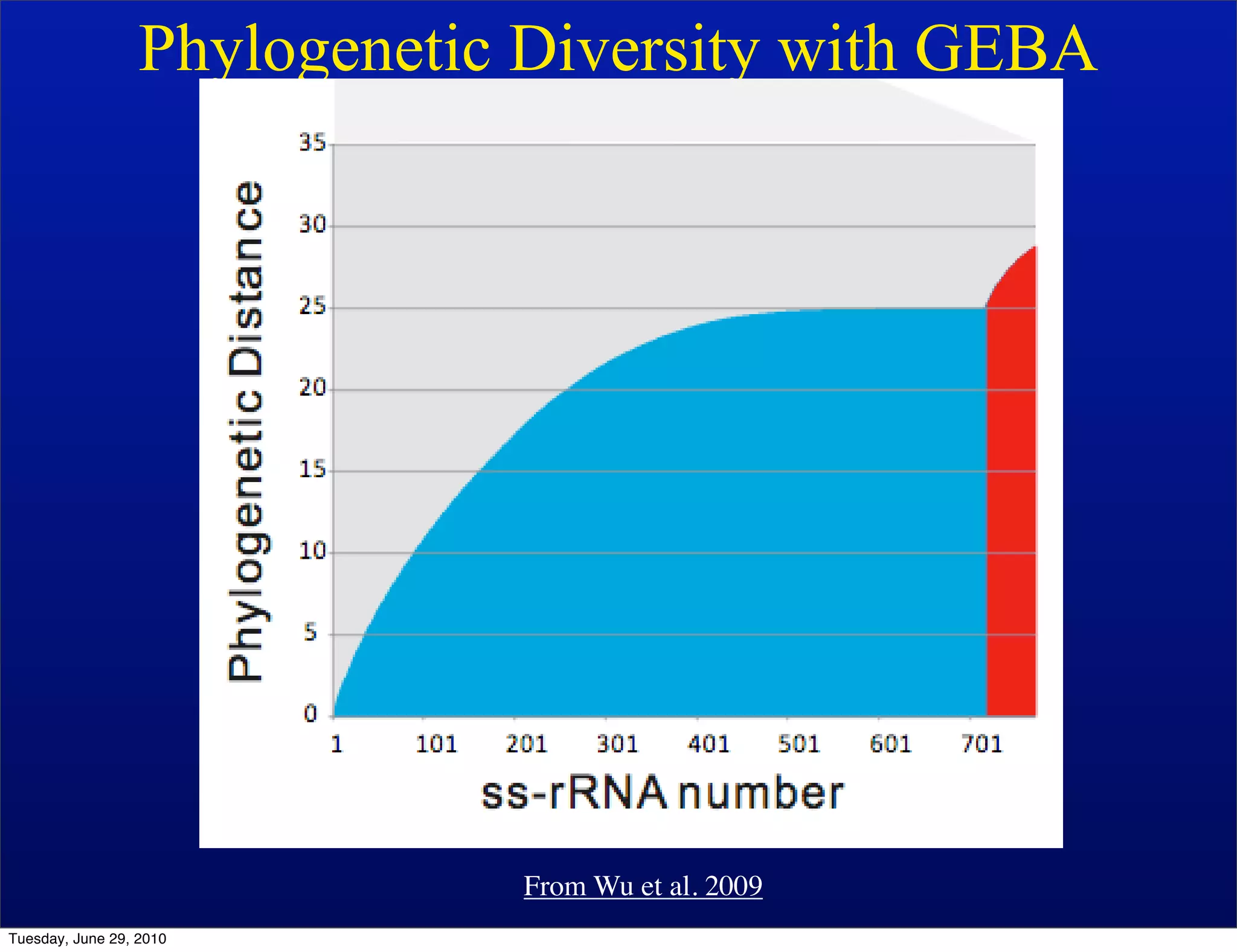
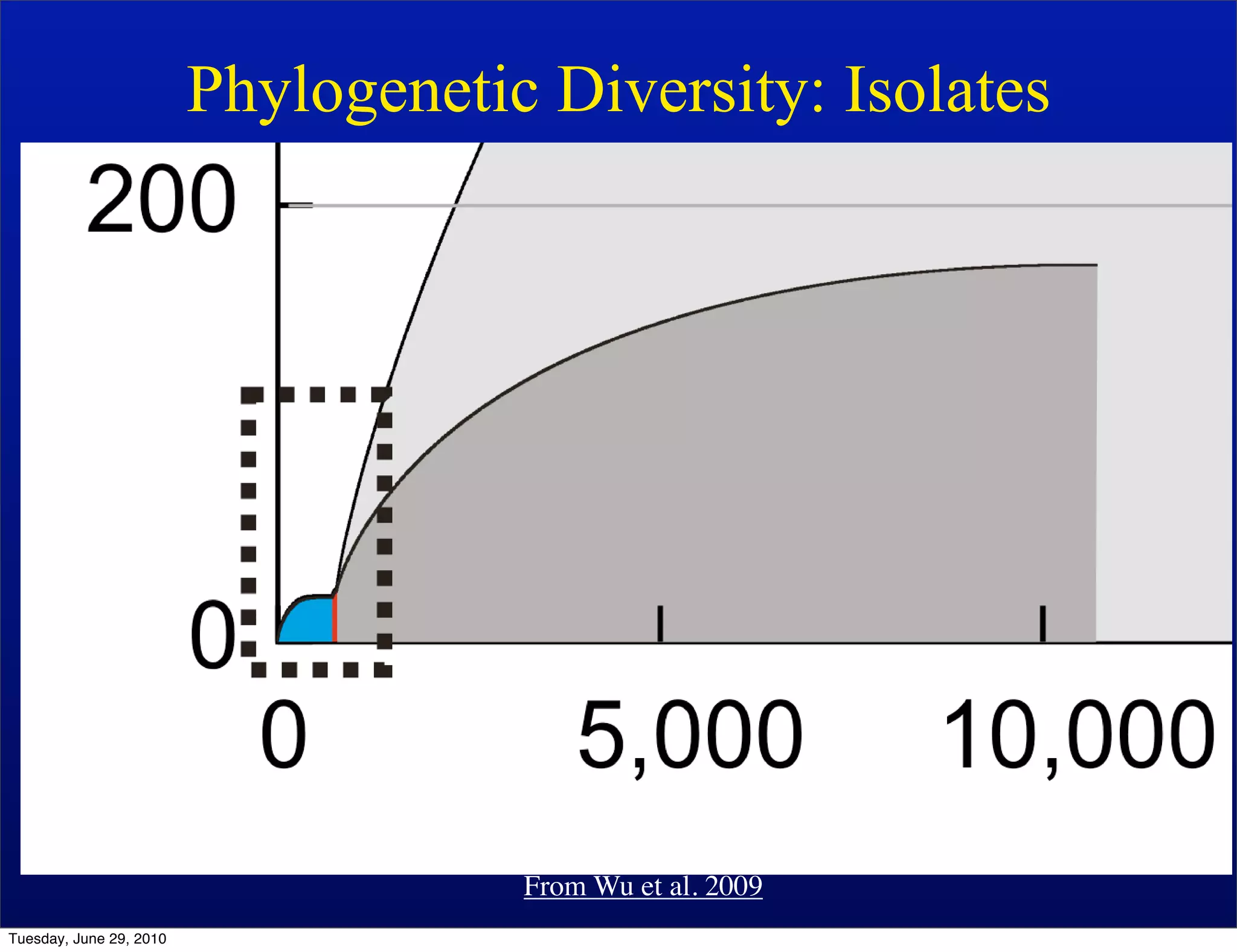
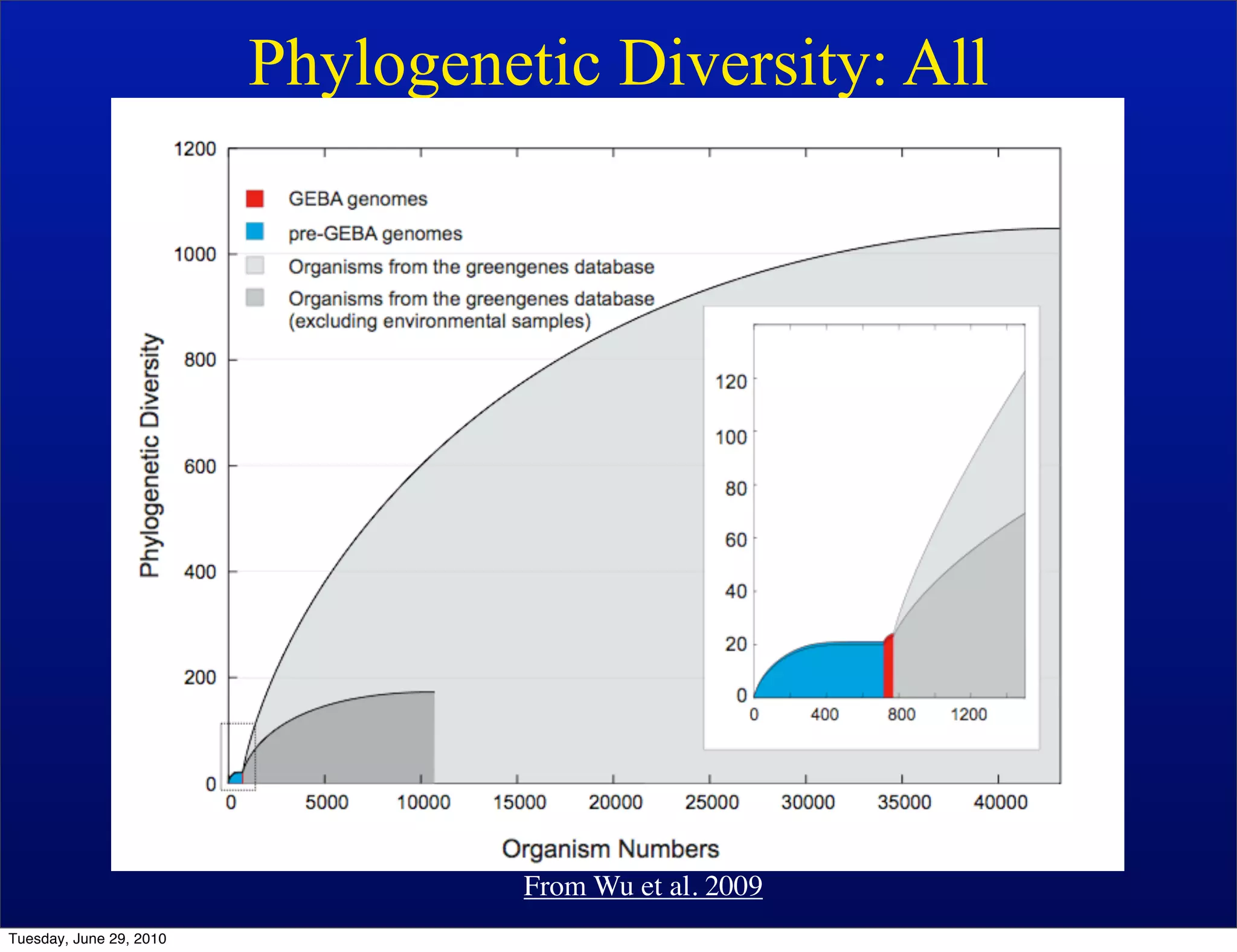
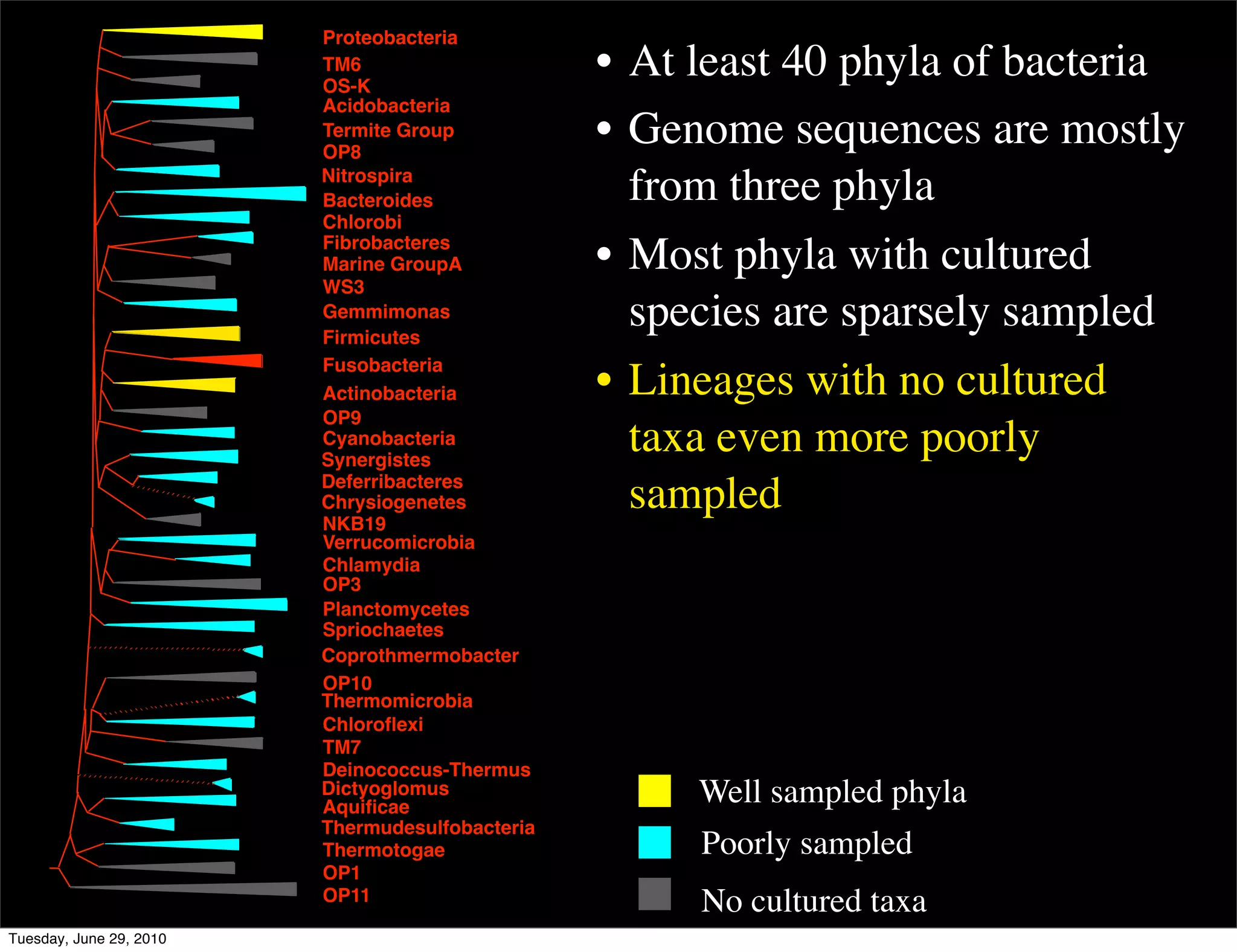
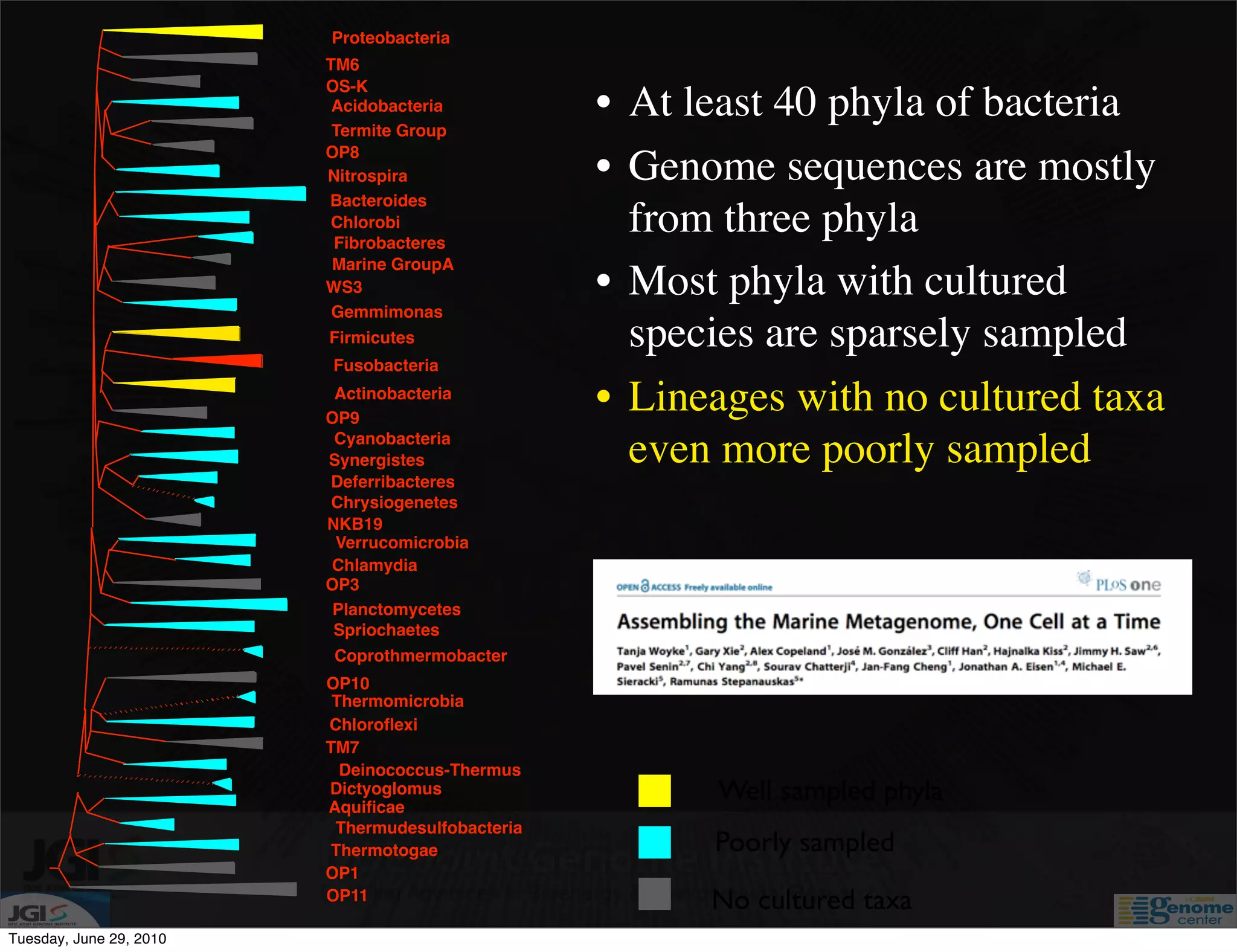
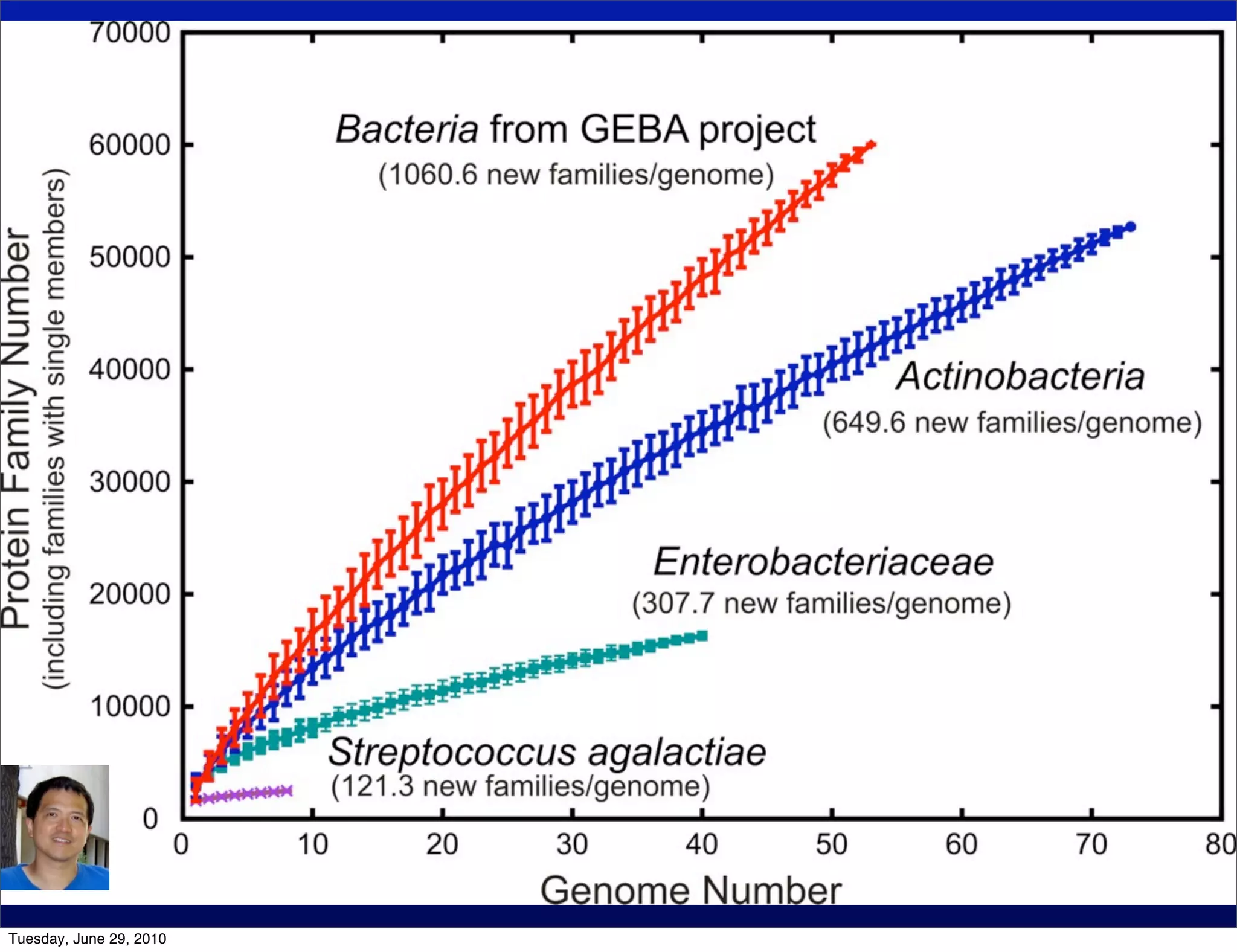
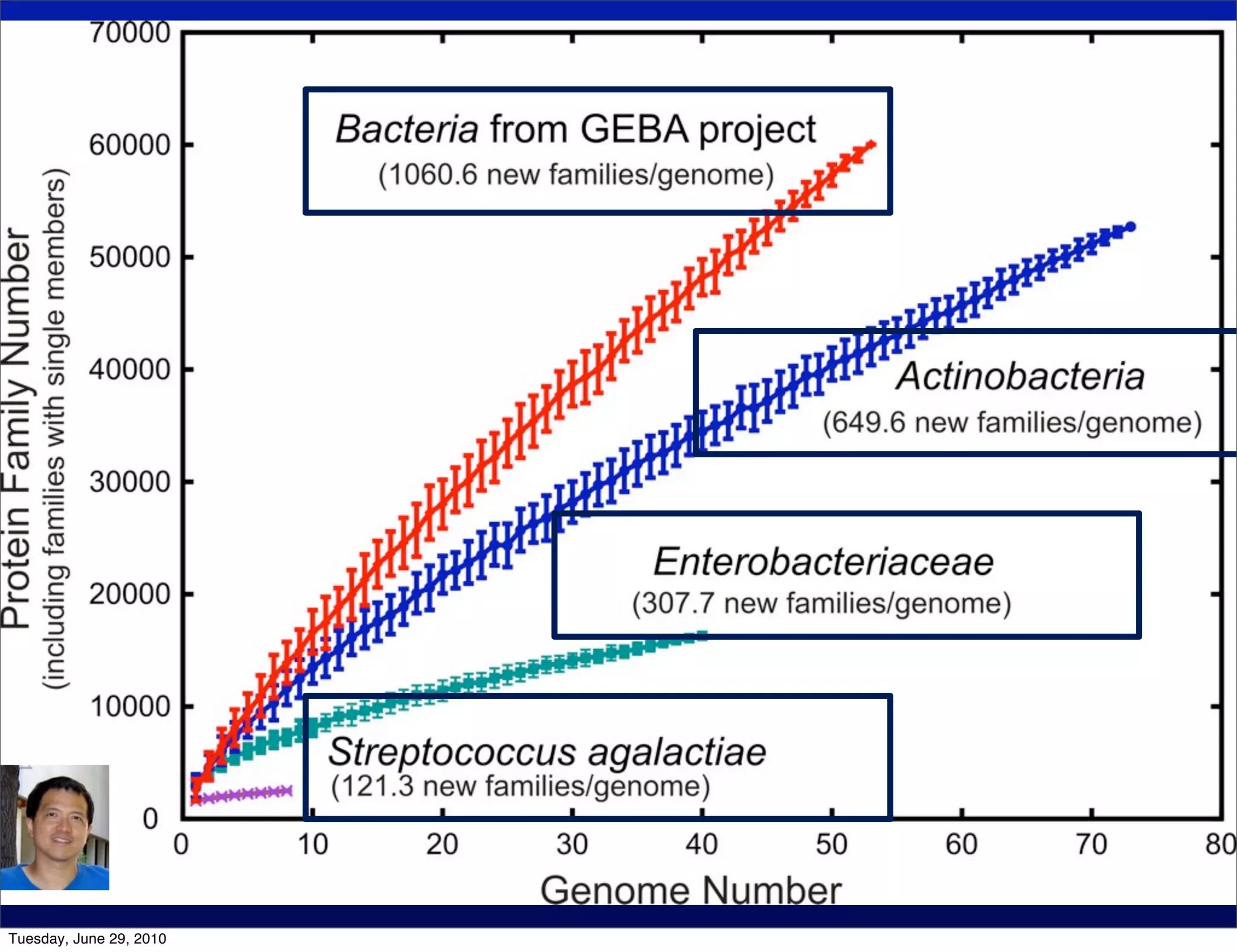
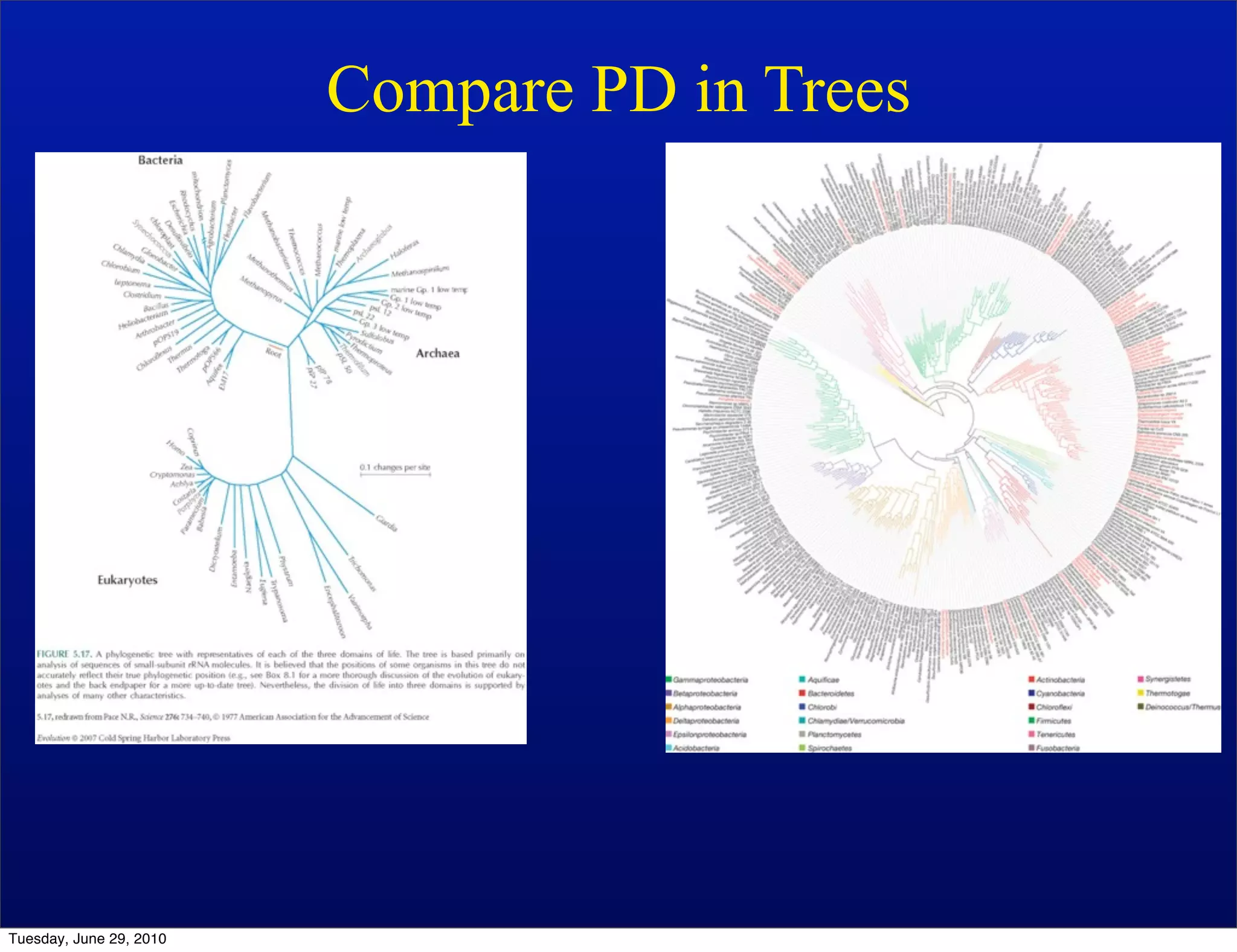
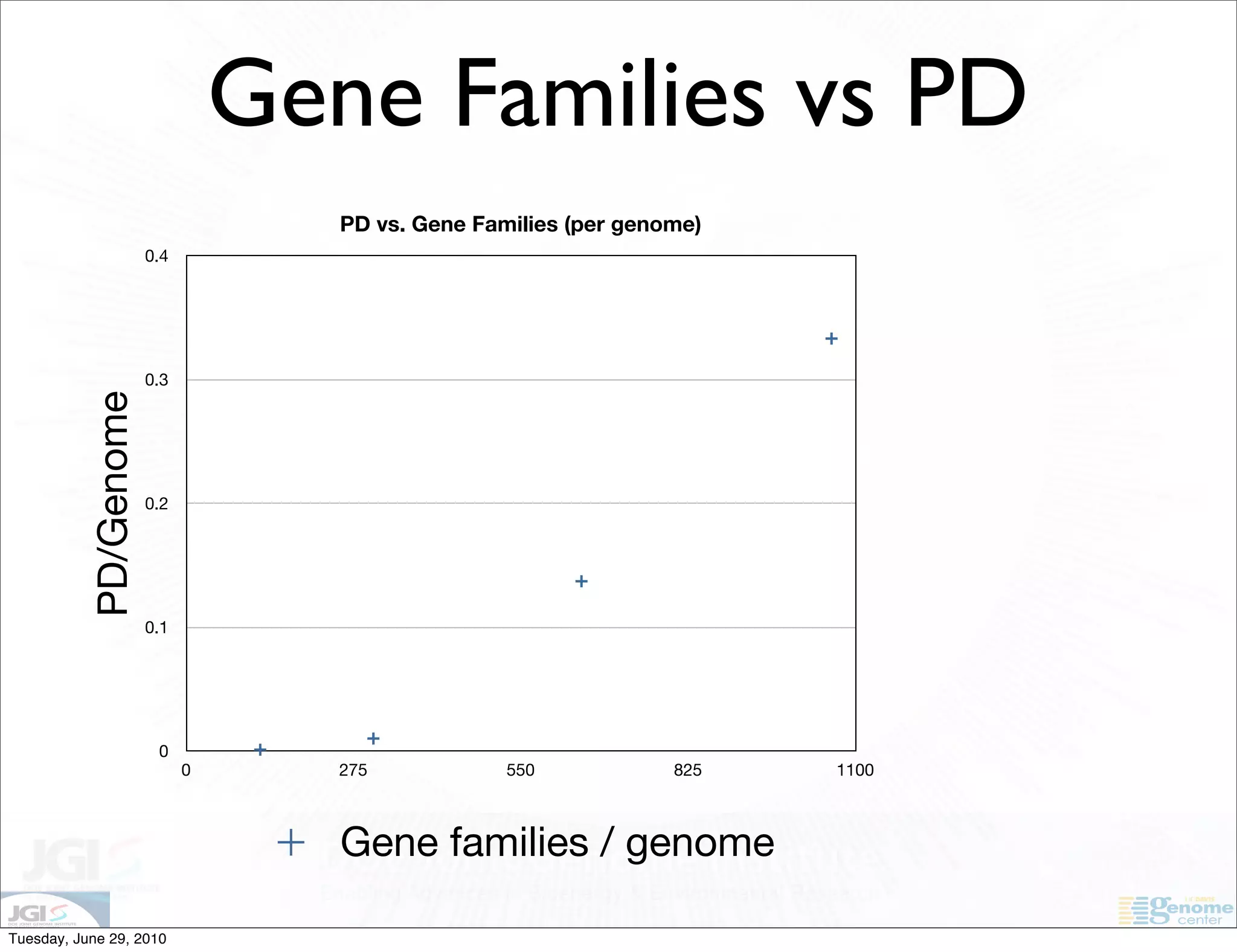
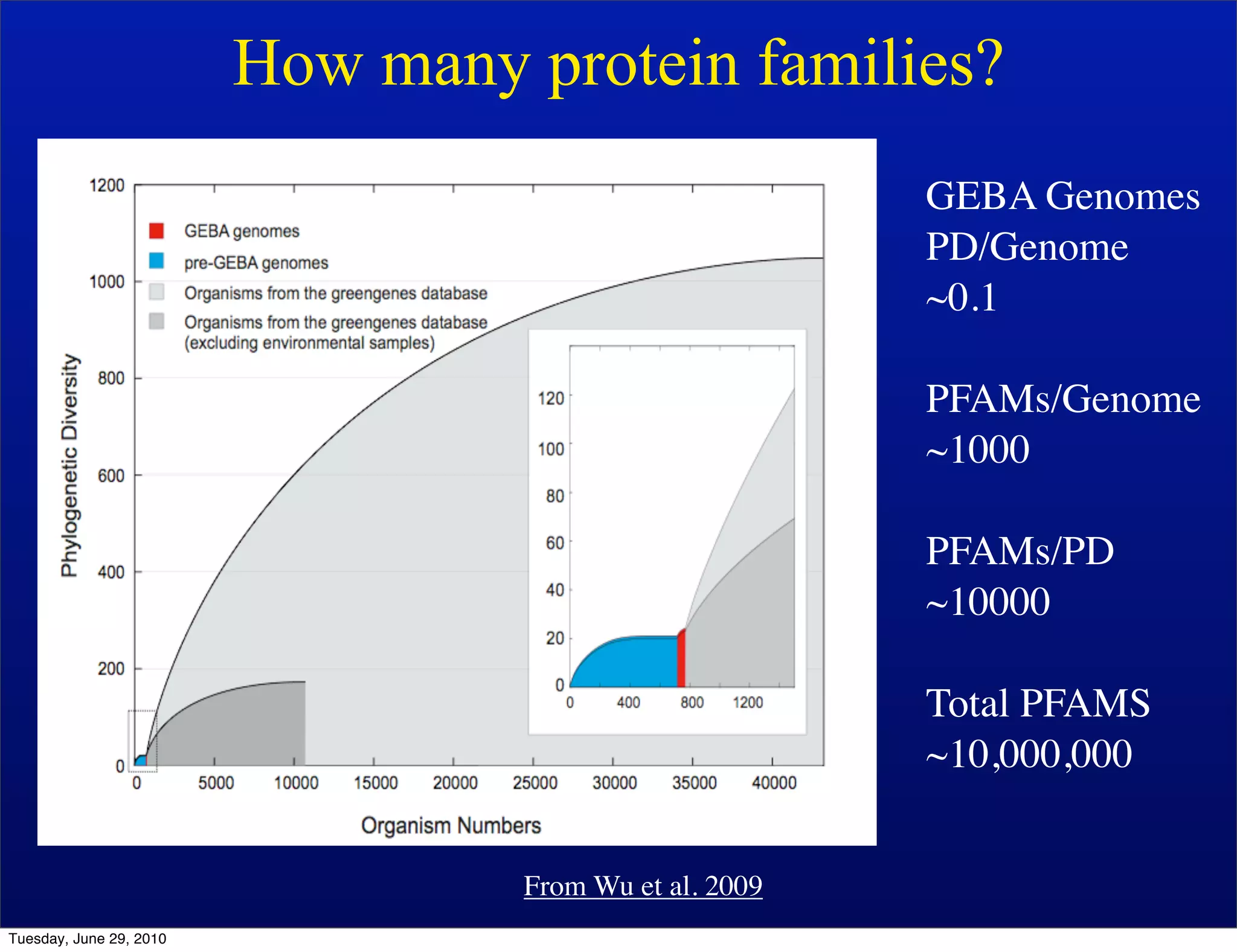
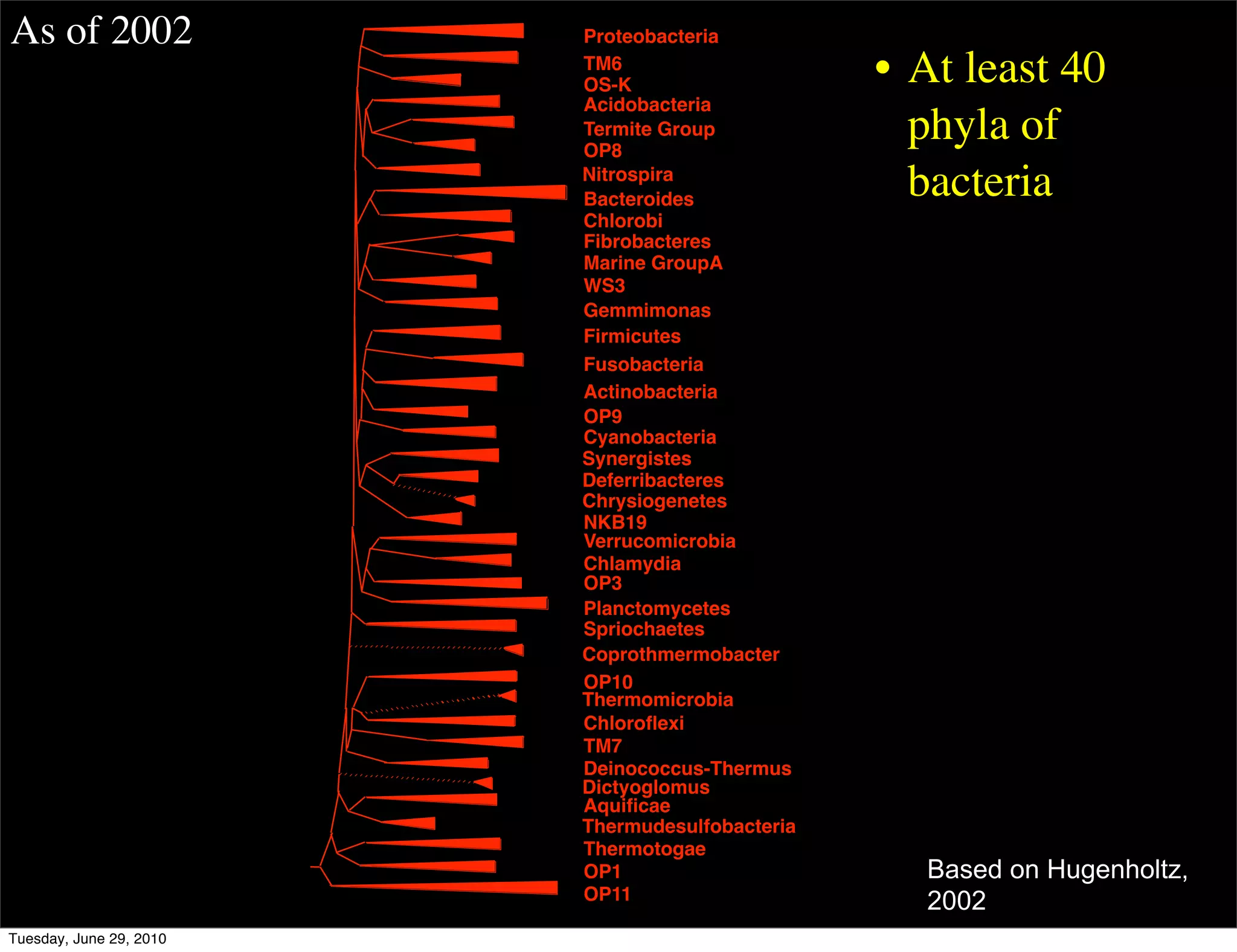
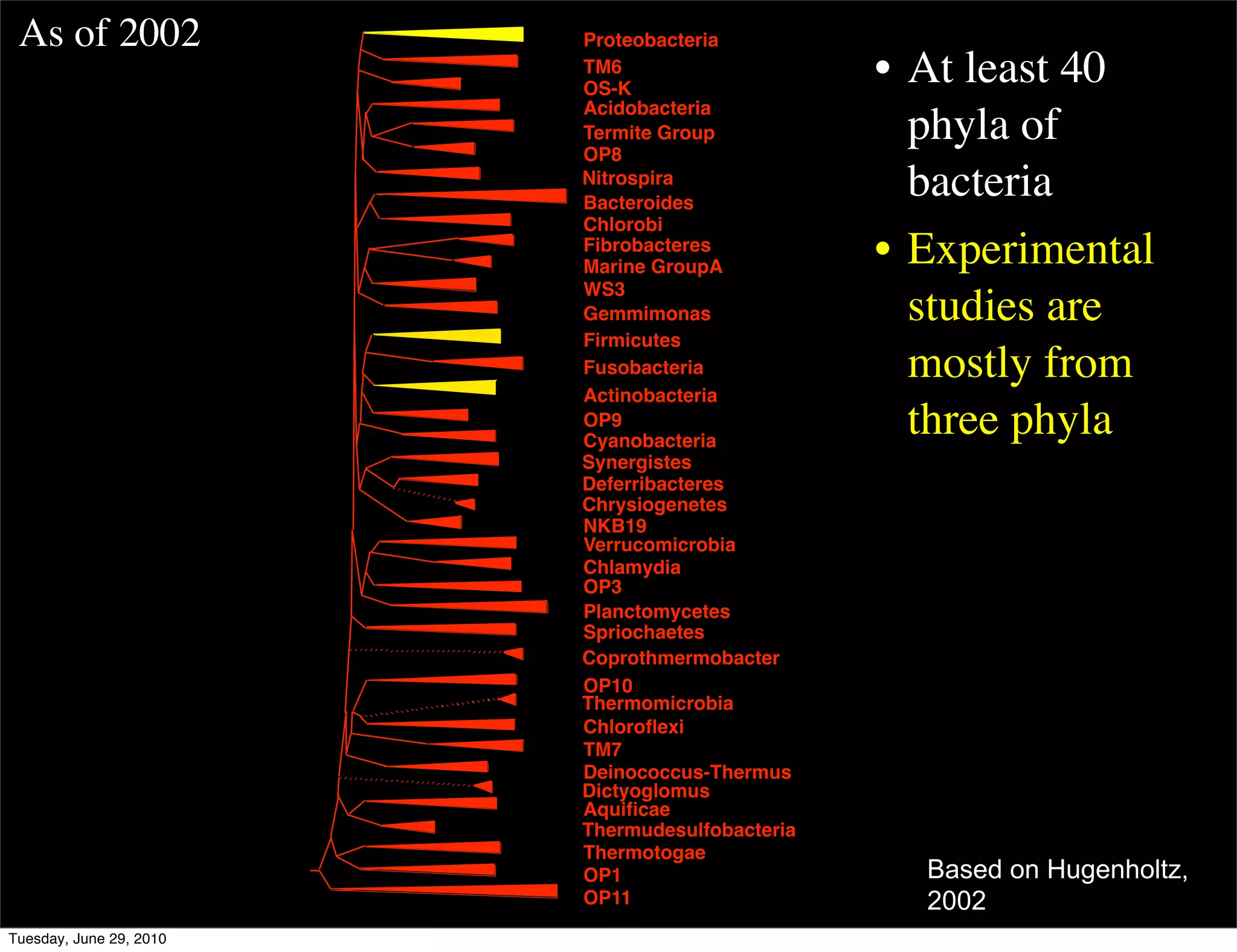
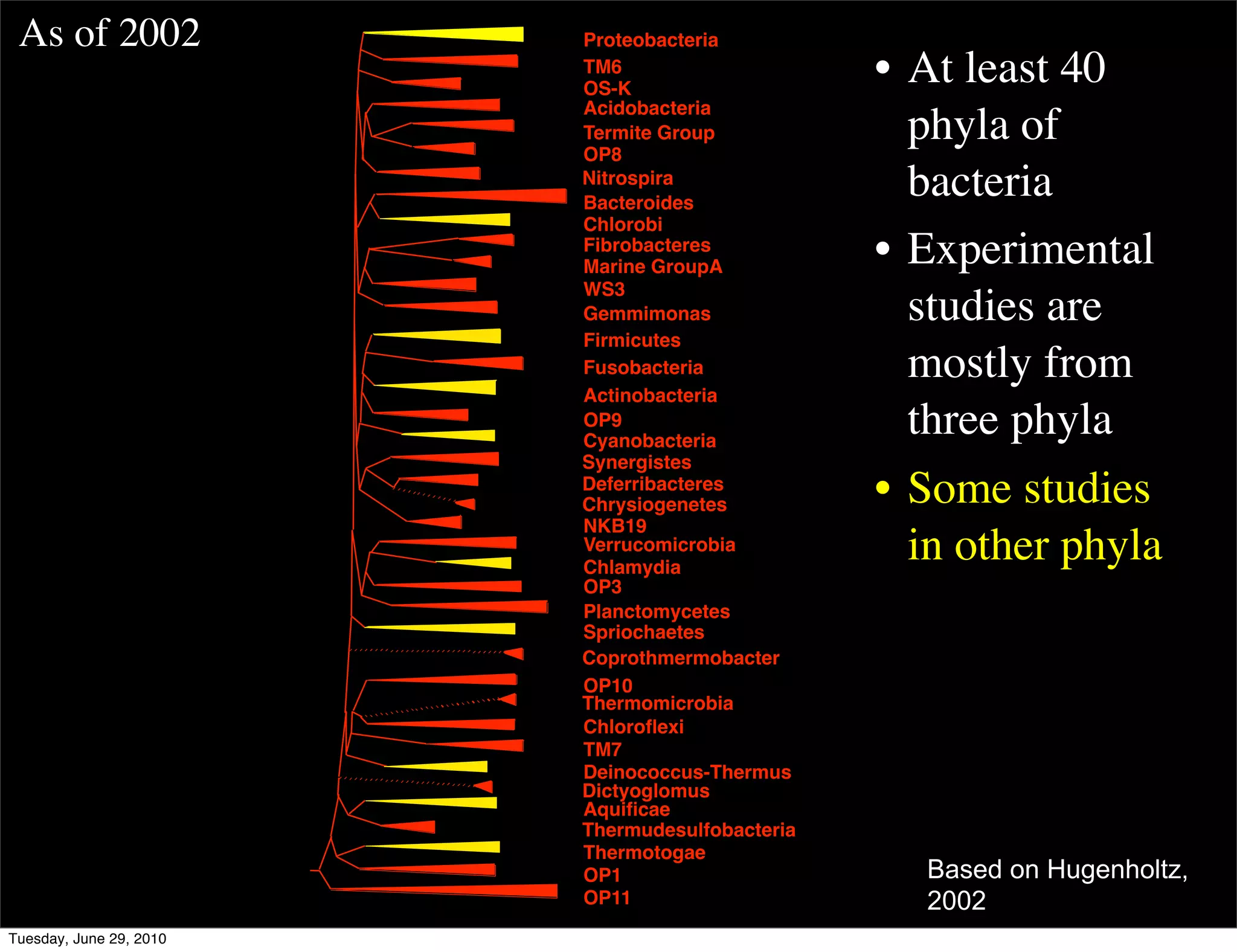
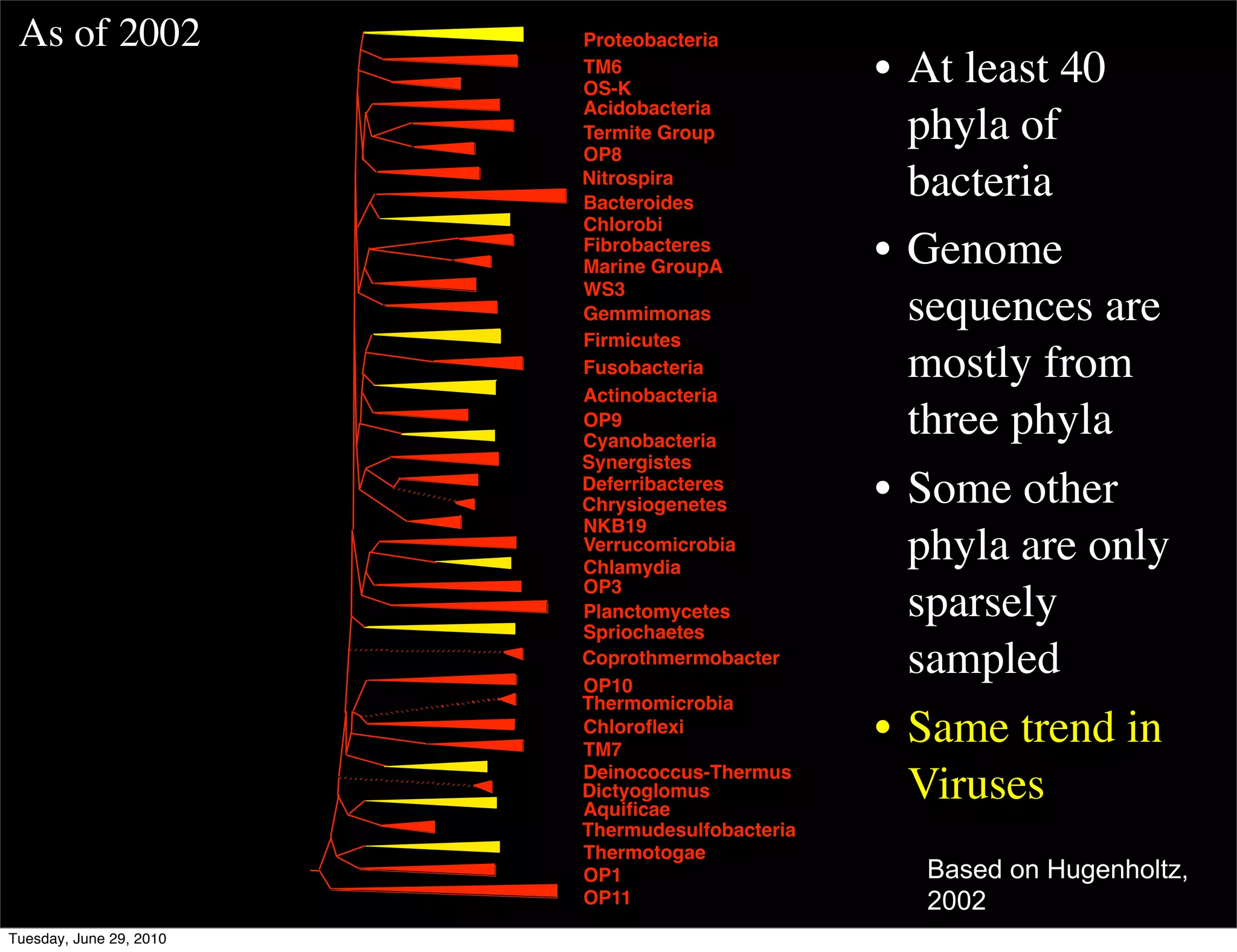
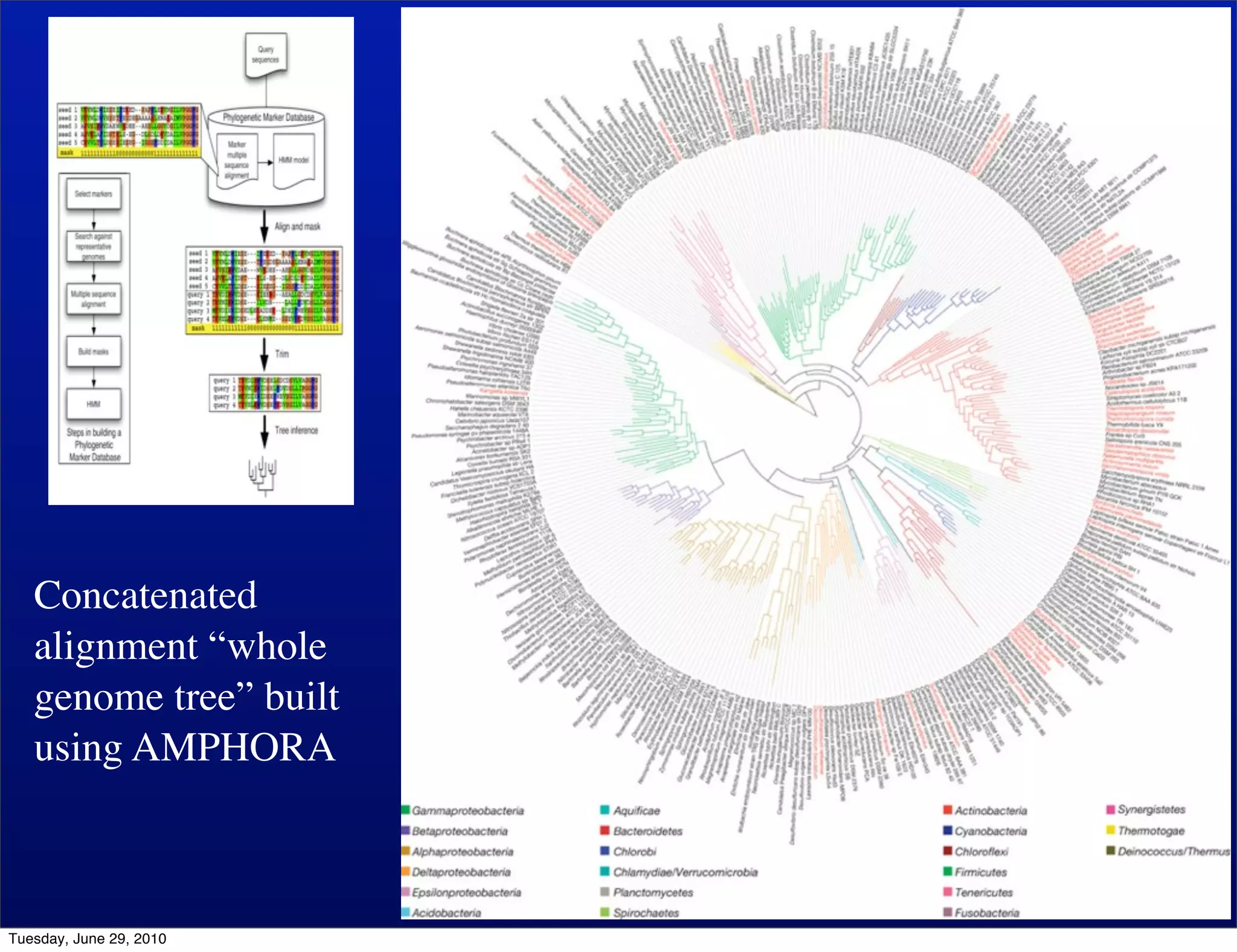
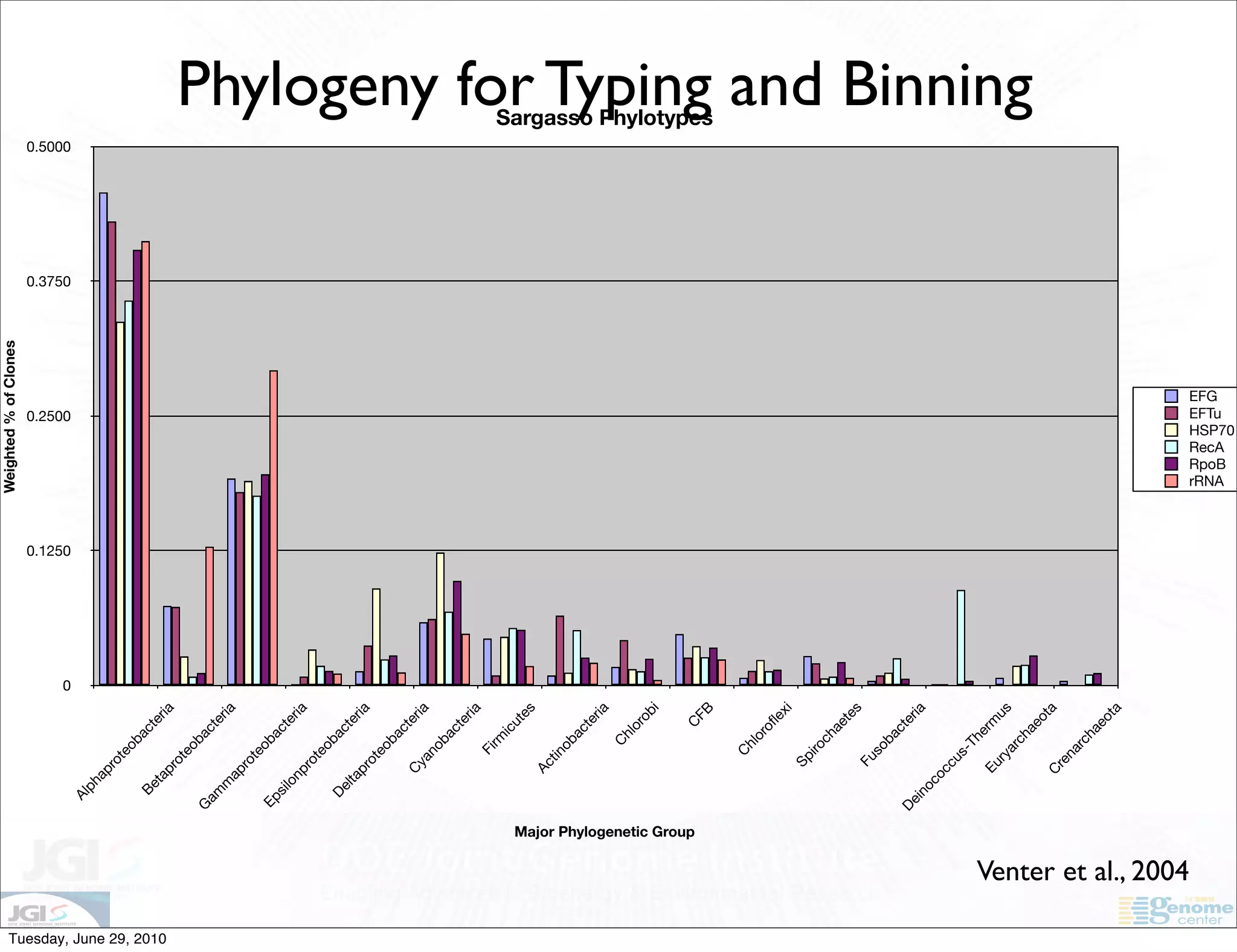
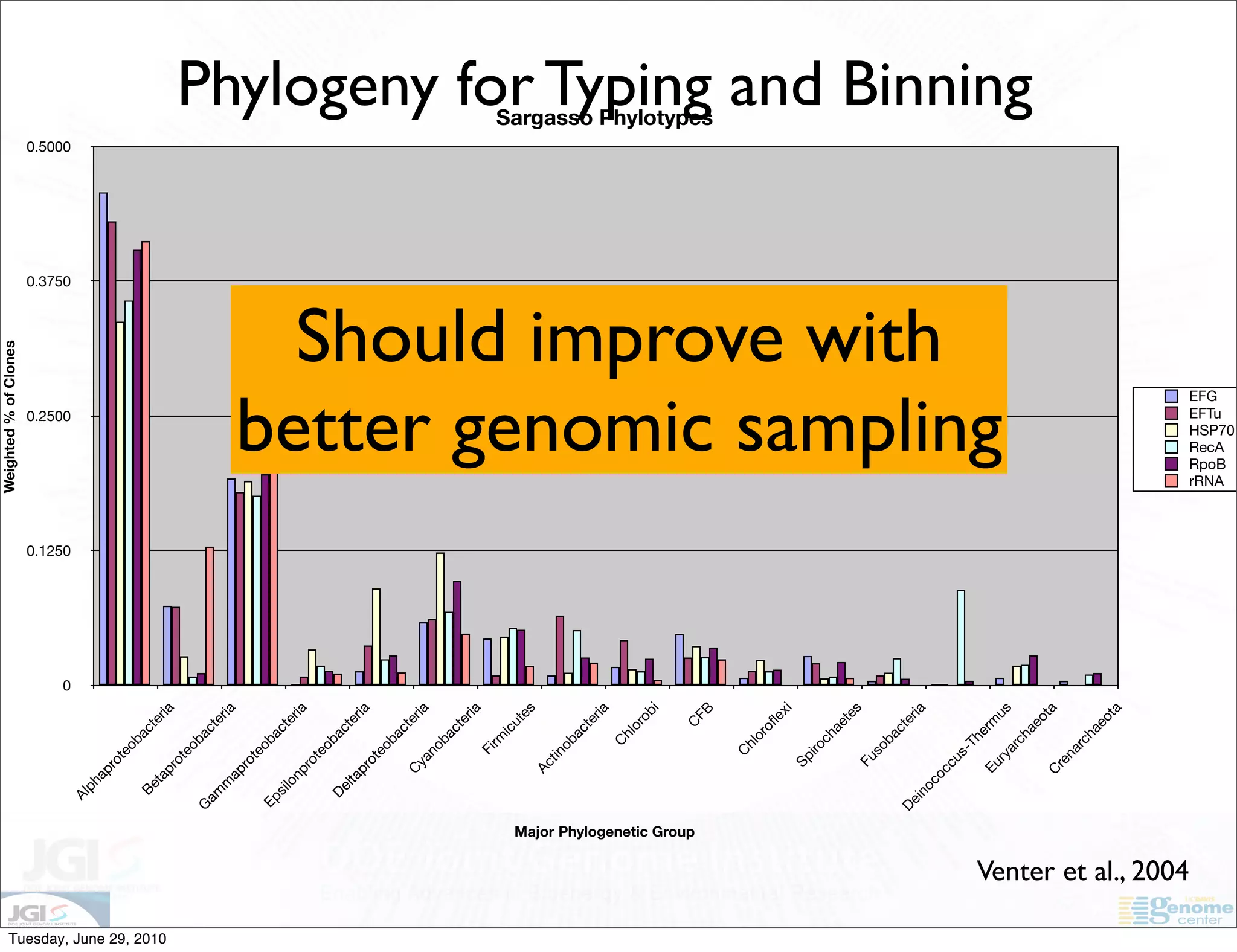
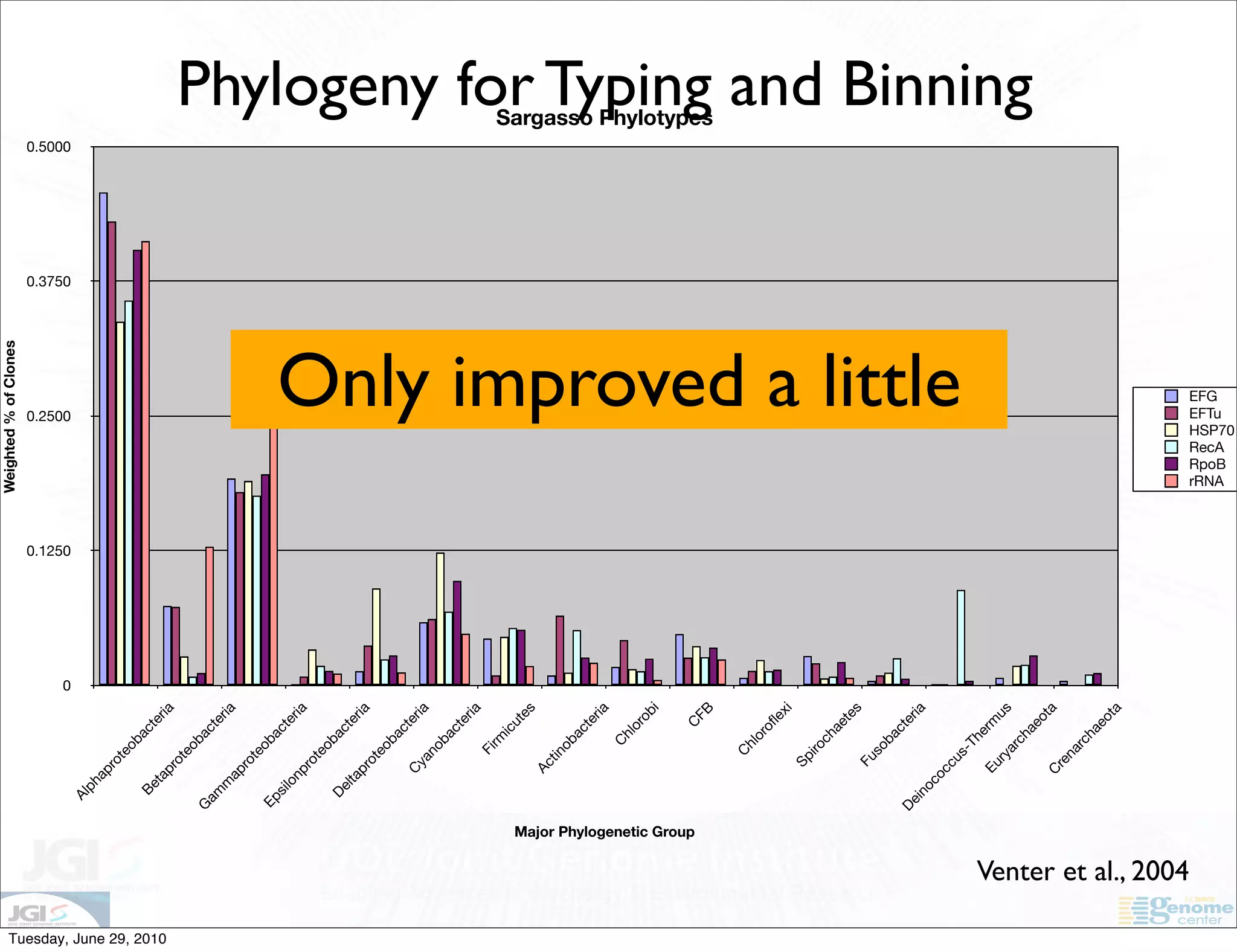
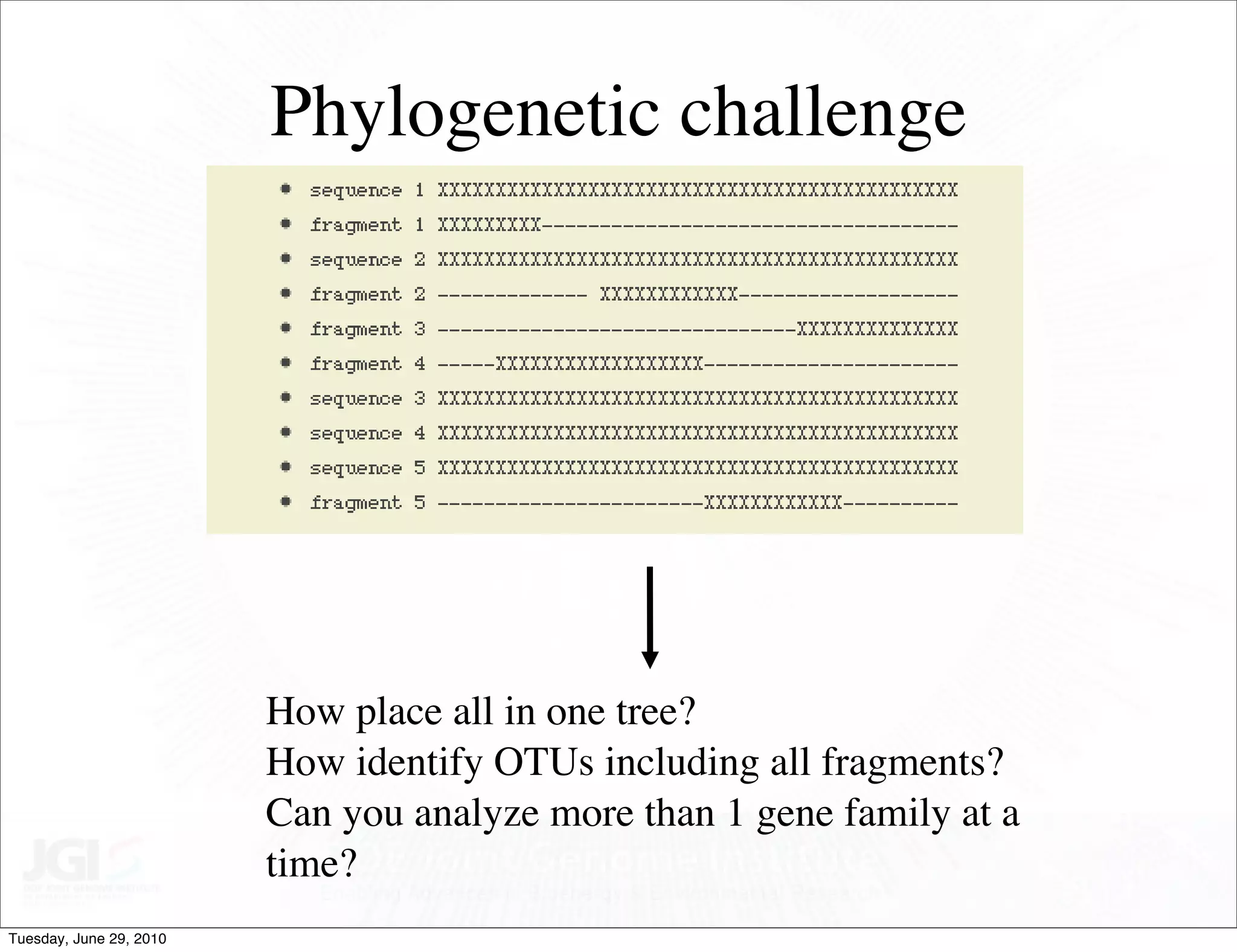
The document discusses the known knowns from the past GEBA project. It notes that as of 2002, genome sequences were mostly from three bacterial phyla, while at least 40 phyla of bacteria were known to exist based on rRNA studies. Some other phyla had only sparse sampling, and the same trend occurred in Archaea and Eukaryotes. More genome sequences were needed from the underrepresented phyla to gain a more comprehensive view of microbial diversity.