Option B Chemistry UV Spectroscopy, Electrophoresis and buffer calculation
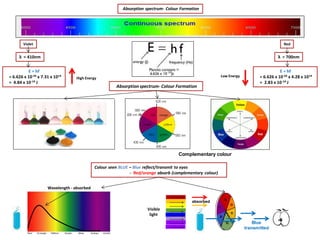
- 1. Absorptionspectrum- Colour Formation Colour seen BLUE – Blue reflect/transmit to eyes - Red/orange absorb (complementary colour) Complementary colour Blue transmitted Wavelength - absorbed Visible light absorbed Absorption spectrum- Colour Formation Violet λ = 410nm E = hf = 6.626 x 10-34 x 7.31 x 1014 = 4.84 x 10-19 J Red λ = 700nm E = hf = 6.626 x 10-34 x 4.28 x 1014 = 2.83 x 10-19 J High Energy Low Energy
- 2. Absorptionspectrum- Colour Formation Colour seen GREEN– GREEN reflect/transmit to eyes - Red/Blue absorb (complementary colour) Complementary colour Green transmitted Wavelength - absorbed Visible light absorbed Violet λ = 410nm E = hf = 6.626 x 10-34 x 7.31 x 1014 = 4.84 x 10-19 J Red λ = 700nm E = hf = 6.626 x 10-34 x 4.28 x 1014 = 2.83 x 10-19 J High Energy Low Energy Absorption spectrum- Colour Formation
- 3. Beer Lambert Law Linear relationshipbet Abs and conc at fix wavelength lc I Io 10logAbsorbance I Io 10log Using UV/vis spectrophotometer ProteinAssay – Amt /Conc protein solution Colour formation – Bind protein with complex dye (Coomassie blue/BSA/Biuret) ↓ Blue complex sol formed ↓ Find wavelength complex absorb – λMax λMax – 595nm– Max absorption at 595nm Protein sol – colourless Complex protein with BSA Add complex dye BSA Colour seen BLUE – Blue reflect/transmit to eyes - Red/orange absorb (complementary colour) Wavelength – λMax 595nm Blue transmitted absorbed Using UV/vis spectrophotometer Monochromator Cuvette Detector Io I Io = Intensity incident light I = Intensity transmitted ε = molar absorption coefficient (constant for absorbing sub I = path length (constant, 1cm) lcAbsorbance concAbsorbance ... ε and I constant
- 4. Beer Lambert Law Linear relationshipbet Abs and Conc at fix wavelength lc I Io 10logAbsorbance I Io 10log Using UV/vis spectrophotometer Monochromator Cuvette Detector Io I lcAbsorbance concAbsorbance ... ε and I constant Absorbance - Unknown protein conc determine by interpolation - Abs unknown is 1.11, by interpolation, conc is 0.75mg/l - Must be diluted , at high conc , deviation from Law interpolation Tube Vol, Protein BSA Vol H2O Vol Buffer Dil Protein conc Abs/ 540nm 1 0.00 2.00 2.00 0.00 0.00 2 0.10 1.90 2.00 0.125 0.34 3 0.30 1.70 2.00 0.375 0.67 4 0.50 1.50 2.00 0.625 1.02 5 0.70 1.30 2.00 0.875 1.35 6 1.00 1.00 2.00 1.25 1.65 Tube 1-6 are diluted protein with complex dye BSA, prepared using initial protein conc of 5.00mg/l Add BSA Add water/buffer Protein – colourless Complex protein/ BSA Diluted protein/BSA (5.00mg/l) 0.125 0.375 0.625 0.875 1.25 0.34 0.67 1.02 1.35 1.65 Std calibration plot, known protein conc vs Abs Unknown – Absorbance = 1.11 Conc = 0.75mg/l interpolation Abs = 1.11 Conc = 0.75 mg/l protein conc
- 5. Beer Lambert Law Linear relationshipbet Abs and Conc at fix wavelength lc I Io 10logAbsorbance I Io 10log Monochromator Cuvette Detector Io I lcAbsorbance concAbsorbance ... ε and I constant protein conc Absorbance interpolation 0.125 0.375 0.625 0.875 1.25 0.34 0.67 1.02 1.35 1.65 Std calibration plot, known protein conc vs Abs Beer’s Lambert Law • Apply for diluted solution • Absorbance α Conc • Absorbance,A = log10 (Io/I) = έlc Absorbance I Io 10log Abs = έlc If έ and l = constant Amt light absorb depend on • έ = Molar extinction compound • c = Conc • l = path length Molar extinctionof compound, έ : • Measure strength of absorption of sub • Higher έ ↑ = Higher ↑ Absorbance • Sub with high έ = effectiveat absorbing light even when low conc is used. Path Length, l: • Longer path length ↑ – Higher ↑ Abs Concentration,c: • Higher conc of analyte – Higher ↑ Abs Abs α Conc, c Using UV/vis spectrophotometer
- 6. Molar absorptivity,έ έ = A/bc = 0.3554 /(2.10 x 7.25 x 10-5) = 2.33 x 103 L mol-1cm-1 Determine conc of unknown using Beer-Lambert Law 7.25 x 10-5M X has absorbance of 0.355 when measured in 2.10 cm cell at wavelength 525nm. Cal Molar absorptivity, έ 100dm3 contaminated water was reduced by boiling to 7.50dm3. Reduced vol was tested, its absorbance is 2.00. Cal conc Pb2+ (mgdm3) in original sample. V = 100 dm3 M = ? V = 7.50 dm3 M = 1.15mg/dm3 Amt bef heating= Amt aft heating Moles bef = Molesaft M x V = M x V M x 100 = 1.15 x 7.50 M = (1.15 x 7.50)/100 M = 0.0863 mgdm-3 Std calibration plot, Abs vs Conc Pb Absorbance 0.3 0.5 0.7 1.1 1.2 Pb conc interpolation Conc Pb2+ 1.15 mgdm-3 Std calibrationcurve, Abs vs Conc Conc unknown (Pb2+) by interpolation Abs = 0.340 Conc = 0.310 1 2 3 Determine unknown conc of Pb2+ using std calibration plot. If unknown sample has Abs 0.34, find conc Pb Abs Conc
- 7. Determine conc of unknown using Beer-Lambert Law V = 5 cm3 M = ? Amt bef dilution= Amt aft dilution Moles bef = Molesaft M x V = M x V M x 5 = 0.38 x 100 M = (0.38 x 100)/5 M = 7.6 mgcm-3 Std calibration plot, Abs vs Conc protein Absorbance o.1 0.2 0.3 0.4 protein conc interpolation Conc protein 0.38 mgcm-3 4 5 cm3 sample protein diluted with buffer to vol of 100 cm3 and analysed with UV. Abs was 1.85 Using std calibration plot, determine conc protein in original sample. V = 100 cm3 M = 0.38 2 cm3 protein was diluted with buffer to vol of 25 cm3 and analysed with UV. Abs was 0.209 Using std calibration plot, determine conc protein in original sample. 5 Conc protein 0.310 mmoldm-3 V = 2 cm3 M = ? V = 25 cm3 M = 0.310 Amt bef dilution= Amt aft dilution Moles bef = Molesaft M x V = M x V M x 2 = 0.310 x 25 M = (0.310 x 25)/2 M = 3.87 mmoldm-3 interpolation
- 8. ChromatographyTechniques • Separation technique of mix into their pure components • Identify sample - mix or pure both quantitatively and qualitatively • Interaction of sub bet 2 phase - Stationary phase and Mobile phase • Separation based on Partitionor Adsorption Chromatography Techniques Separation analysis Paper Chromatography Thin Layer Chromatography Adsorption Chromatography Chromatography Partition Chromatography Column Chromatography Partition Chromatography • Component distribute bet TWO immisible liquid phase • Depend on relative solubility bet TWO phase • Solutes bond to stationary phase or mobile phase Adsorption Chromatography • Component adsorb on solid stationaryphase • Depend on polarity of stationary,mobile phase and solutes • Stationary phase is polar – polar solutes adsorb strongly • Stationary phase is non polar – non polar solutes adsorb strongly • Mobile phase is polar – polar solutes stay in mobile phase • Mobile phase is non polar – non polar solutes stay in mobile phase Application • Detectionamino acids in mix • Diff dyes in food colouring • Separation plant pigments Y adsorb strongly Application Collection of sample of pigments X adsorb strongly Application • Detectionamino acids in mix • Diff dyes in food colouring • Separation plant pigments X adsorb weakly
- 9. Chromatography Partition Chromatography • components distribute bet 2 immisible liquid phase • relative solubility in 2 phase • bond strongly to mobile phase – move faster Adsorption Chromatography • Components adsorp on solid stationaryphase Stationary phase has layer of liq Mobile liq phase containing X and Y X X X Y Y Y Stationary Liquid phase Partition –distribution solute X and Y bet 2 liq phase • X more soluble in mobile phase (move with mobile liq phase) • Y less soluble in mobile phase (stay on stationary liq phase) Stationary phase has layer of liq Y Y Y Y X XX XX XMobile Iiq phase containing X Stationary phase • solid • AI2O3 • SiO2 O- O- O- O- O- Adsorption – solute X and Y adsorb temporary on solid • Y adsorb strongly on solid phase, eluted slower • X in liq mobile phase, eluted faster Y Y Y Y Mobile liq phase containing X and Y X X X O- O- O-Y Y Y Y X X XX Mobile liq phase containing X Y adsorb strongly X X X X Y Y Y Y Separation of X and Y X X X Y Y Y Chromatography Techniques
- 10. Paper Chromatography Partition chromatography • Distribution solute bet both liquid phase • Depend on relative solubility Aqueous liq phase on surface of stationary phase (paper) Mobile liq phase - solvent Solvent move by capillary action Stationary phase - Cellulose paper • absorb water on its surface Mobile Liquid phase with solute X and Y Y Y Y X X Chromatography Techniques Componentseparatedidentifiedusing Rf value • Rf = Retention factorfor given eluent. • Measured distance from original spot to centre of particularcomponent to solvent front solventbyceDis solutebyceDis factortention ....tan ....tan .Re Rf green spot = (3/12) = 0.25 Rf blue spot = 6/12 = 0.5 Separation using TLC TLC techniquesstep by step Column Chromatography Column separation
- 11. Electrophoresis Amino acid Zwitterion (Electrically neutral) Amino acid with isoelectric point Isoelectric point pH when amino acid is electrically neutral Isoelectric point alanine = 6.02 (Ave of pKa) Separation amino acid based on charges using electric field At pH 6.02 Alanine is electrically neutral (Zwitterion) pH = isoelectric Alanine ( Neutral) pH < isoelectric Alanine (+ve) pH > isoelectric Alanine (- ve)
- 12. Electrophoresis Isoelectric point pH when amino acid is electrically neutral Isoelectric point alanine = 6.02 (Ave of pKa) Separation amino acid based on charges using electric field At pH 6.02 Alanine is electrically neutral (Zwitterion) pH = isoelectric Alanine (Neutral) pH < isoelectric Alanine (+ve) pH > isoelectric Alanine (-ve) Mix of amino acid (ala, arg, isoleu, asp acid) Spot at center on gel (polyacrylamide) Buffer, pH 6 added. Electric field applied Amino acid separate based on charges. Ninhydrin applied to identified spots. Separation based on charges pH = iso point = No movement (zwitterion)/ electrically neutral pH < iso point = + ve charge (cation) = move to – ve (cathode) pH > iso point = - ve charge (anion) = move to +ve (anode) pH = 6 + ve - ve
- 13. Electrophoresis Isoelectric point pH when amino acid is electrically neutral Isoelectric point alanine = 6.02 (Ave of pKa) Separation amino acid based on charges using electric field pH = isoelectric Alanine (Neutral) pH < isoelectric Alanine (+ve) pH > isoelectric Alanine (-ve) + H+ Acidic (pH < pI) Cation/zwitterion - H+ + H+ - H+ Cation (conjugateacid) Zwitterion (conjugatebase) + H+ + H+ Zwitterion (conjugateacid) Anion (conjugatebase) Alkaline(pH > pI) Zwitterion/anion
- 14. Cation (+) Zwitterion Anion (-) + H+ Acidic (pH < pI) Cation/zwitterion - H+ + H+ - H+ Cation (conjugateacid) Zwitterion (conjugatebase) + H+ + H+ Zwitterion (conjugateacid) Anion (conjugatebase) Alkaline(pH > pI) Zwitterion/anion Acidic Base Buffer Calculation ][ ][ log1 cation zwitterion pKpH ][ ][ log2 zwitterion anion pKpH Find pH 0.8M zwitterionand 0.2M anionic form serine + H+ + H+ - H+ - H+ Amino acid pK1 pK2 Isoelectric Serine 2.2 9.1 5.7 Zwitterion (conjugate acid) Anion (conjugate base) 5.8 8.0 2.0 log1.9 ][ ][ log2 pH zwitterion anion pKpH
- 15. At pH 7, alanine contain zwitterion and anionic form i. Deduce structuralformula ii. State eqn of buffer when small amt acid and base added Zwitterion (conjugate acid) Anion (conjugate base) Alkaline (pH > pI) Zwitterion/anion Zwitterion (conjugate acid) Anion (conjugate base) + OH- + H+ Acid addedBase added 2 1
- 16. Zwitterion (conjugate base) Cation (conjugate acid) Acidic (pH < pI) Cation/zwitterion Amino acid pK1 pK2 Isoelectric Glycine 2.3 9.6 6.0 Amino acid prepared by mixing 0.6 dm3, 0.2M HCI and 0.4dm3, 0.5M glycine Find pH original buffer n (HCI) = 0.6 x 0.2 = 0.12 mol n (glycine) = 0.4 x 0.5 = 0.20 mol Total vol = 1 dm3 Conc (HCI) = Mol/Vol = 0.12/1 = 0.12M Conc (glycine) = Mol/Vol = 0.20M ][ ][ log1 cation zwitterion pKpH 12.2 ]12.0[ ]08.0[ log3.2 pH pH 2
- 17. ][ ][ log1 cation zwitterion pKpH Zwitterion (conjugate base) Cation (conjugate acid) Acidic (pH < pI) Cation/zwitterion Amino acid pK1 pK2 Isoelectric Glycine 2.3 9.6 6.0 Amino acid prepared by mixing 0.4 mol zwitterion and 0.16mol cationic glycine in 1 dm3 Find pH of buffer Total vol = 1 dm3 Conc (cation) = Mol/Vol = 0.16/1 = 0.16M Conc (zwitterion) = Mol/Vol = 0.4/1 = 0.4M ][ ][ log1 cation zwitterion pKpH 7.2 ]16.0[ ]4.0[ log3.2 pH pH Find pH buffer when - 0.025M of NaOH added to buffer 3 4 0.16 0.025 0.4 - 0.025 - 0.025 + 0.025 0.135 0.425 OH- - -COOH + Conc (zwitterion) = Mol/Vol = 0.4/1 = 0.4M Conc (cation) = Mol/Vol = 0.16/1 = 0.16M 8.2 ]135.0[ ]425.0[ log3.2 pH pH
- 18. NH3 ↔ NH4 + Buffer Solution Acid part Neutralize each other Salt part Base part - NH3(weakbase) + NH4CI (salt) - NH3 + H2O ↔ NH4 + + OH− → NH3 neutraliseadded H+ - NH4CI → NH4 + + CI− → NH4 + neutralise added OH− - Effectivebuffer equal amt weak base NH3 and conjugate acid NH4 + Acidic Buffer Basic Buffer Resist a change in pH when small amt acid/base added. CH3COOH + H2O ↔ CH3COO- + H3O+ Acidic Buffer - weak acid and its salt/conjugatebase CH3COOH ↔ CH3COO- Conjugate acid base pair CH3COOH CH3COO- Weak Acid Conjugate Base BUFFER Dissociate fully HCOOCHCOOHCH 33 COOHCH3 COONaCH3 NaCOOCHCOONaCH 33 Dissociate partially - CH3COOH (weakacid) + CH3COONa (salt) - CH3COOH ↔ CH3COO- + H+ → CH3COOH neutraliseadded OH− - CH3COONa → CH3COO- + Na+ → CH3COO- neutraliseadded H+ - Effectivebuffer equal amt weak acid CH3COOH and base CH3COO- COOHCH3 COOCH3 BUFFER Add acid H+Add alkaline OH- Neutralize each other Basic buffer - weak base and its salt/conjugateacid OHNHOHNH 423 NH3 + H2O ↔ NH4 + + OH- NH3 Weak Base NH4 + Conjugate acid CINH43NH BUFFER Conjugate acid base pair Add acid H+ Add alkaline OH- Neutralize each other Neutralize each other Dissociate partially CINHCINH 44 3NH 4NH Base part Salt part Acid part Dissociate fully BUFFER
- 19. How to prepare acidic/ basic buffer Acid Dissociationconstant CH3COOH + H2O ↔ CH3COO- + H3O+ Ka = (CH3COO- ) (H3O+ ) (CH3COOH) -lgKa = -lgH+ -lg (CH3COO-) (CH3COOH) -lgH+ = -lg Ka + lg (CH3COO-) (CH3COOH) pH = pKa + lg (CH3COO-) (CH3COOH) Acidic Buffer Formula • Mix Weak acid + Salt/Conjugate base • CH3COOH ↔ CH3COO- + H+ (dissociate partially) • CH3COONa → CH3COO- + Na+ (dissociate fully) Basic Buffer Formula • Mix Weak base + Salt/Conjugate acid • NH3 + H2O ↔ NH4 + + OH_ (dissociate partially) • NH4CI → NH4 + + CI_ (dissociate fully) pH = pKa - lg (acid) (salt) pH = pKa + lg (salt) (acid) Base Dissociationconstant NH3 + H2O ↔ NH4 + + OH- Kb = (NH4 + ) (OH- ) (NH3) -lgKb = -lgOH- -lg (NH4 +) (NH3) -lgOH- = -lgKb + lg (NH4 +) (NH3) pOH = pKb + lg (NH4 +) (NH3) pOH = pKb + lg (salt) (base) pOH = pKb - lg (base) (salt) Basic BufferAcidic Buffer salt salt acid base Henderson Hasselbalch Eqn multiply -lg both sides Henderson Hasselbalch Eqn
- 20. Acidic BufferCalculation How much 0.10M butanoic acid and solid potassium butanoate neededto make 1.0 dm3, pH 5.00 buffer solution. State assumption used. pKa acid = 4.83 Need 0.15 mol in 1 dm3 Mass salt = mol x RMM = 0.15 x 126.12 = 19 g salt Click here video Khan Academy Find pH buffer made with 0.20 mol CH3COONa(salt) in 500cm3 of 0.10M CH3COOH(acid) Ka = 1.8 x 10-5 Find pH buffer - adding 25 ml, 0.10M CH3COOH(acid) 25ml, 0.10M CH3COONa(salt) Ka = 1.8 x 10-5 1st method (formula) 1 Convert Ka to pKa 2nd method (Ka) 2 1st method (formula) 3 1st method (formula) Molar mass salt = 126.12 gmol-1 2nd method (Ka) Click here explanation from chem guide 74.4 ]05.0[ ]05.0[ lg74.4 ][ ][ lg pH pH salt acid pKpH a 74.4 )108.1lg( )lg( 108.1 05.0 ))(05.0( 108.1 )( ))(( 5 5 5 3 3 pH pH HpH H H COOHCH HCOOCH Ka 3 /15.0][ ][ ]10.0[ lg83.400.5 ][ ][ lg dmmolsalt salt salt acid pKpH a 74.4 )108.1lg( lg 108.1 5 5 a a aa a pK pK KpK K 35.5 105.4][ 10.0 ))(40.0( 108.1 )( ))(( 6 5 3 3 pH MH H COOHCH HCOOCH Ka 35.5 ]40.0[ ]10.0[ lg74.4 ][ ][ lg pH pH salt acid pKpH a 3 /40.0 50.0 20.0 dmmolconc conc vol mol conc Conc salt Equal vol and conc Ratio acid/salt = 1 Assumption used • [butanoic acid]eq = [salt] used • [acid]eq = [acid] used • No vol change during mixing
- 21. Acidic BufferCalculation Find pH buffer - 0.20 mol CH3COONa(salt) add to 0.5dm3, 0.10M CH3COOH(acid) Ka = 1.8 x 10-5 Conc CH3COO- =Moles/Vol = 0.20/0.5 = 0.40M Click here video0 Khan Academy Find conc CH3COONa(salt) added to 1.0dm3 of 1.0M CH3COOH(acid) Ka = 1.8 x 10-5, pKa = 4.74 , pH 4.5 Find pH buffer - 0.10M CH3COOH(acid), 0.25M CH3COONa(salt) Ka = 1.8 x 10-5 1st method (formula) 4 Convert Ka to pKa 2nd method (Ka) 5 1st method (formula) Convert Ka to pKa 2nd method (Ka) 6 1st method (formula) Conc salt 2nd method (Ka) Click here explanation from chem guide 14.5 ]25.0[ ]10.0[ lg74.4 ][ ][ lg pH pH salt acid pKpH a 14.5 )102.7lg( )lg( 102.7 10.0 ))(25.0( 108.1 )( ))(( 6 6 5 3 3 pH pH HpH H H COOHCH HCOOCH Ka 34.5 ]40.0[ ]10.0[ lg74.4 ][ ][ lg pH pH salt acid pKpH a 74.4 )108.1lg( lg 108.1 5 5 a a aa a pK pK KpK K 74.4 )108.1lg( lg 108.1 5 5 a a aa a pK pK KpK K MCOOCH COOCH COOHCH HCOOCH Ka 0578.0 0.1 )1016.3)(( 108.1 )( ))(( 3 5 35 3 3 Msalt salt salt salt acid pKpH a 0578.0][ 24.0 ][ ]0.1[ lg ][ ]0.1[ lg74.45.4 ][ ][ lg 34.5 )105.4lg( )lg( 105.4 10.0 ))(40.0( 108.1 )( ))(( 6 6 5 3 3 pH pH HpH H H COOHCH HCOOCH Ka 5 1016.3 )lg(5.4 )lg( H H HpH Conc [H+]
- 22. Find pH buffer - 0.50M NH3 (base), 0.32M NH4CI (salt) Kb = 1.8 x 10-5 Basic Buffer Calculation Find pH buffer - 4.28g NH4CI (salt) add to 0.25dm3, 0.50NH3(base) Kb = 1.8 x 10-5 Mole NH4CI = mass/RMM = 4.28 / 53.5 = 0.08 mol Conc NH4CI = moles/vol = 0.08/0.25 = 0.32M 7 1st method (formula) 2nd method (Kb) 1st method (formula) 8 2nd method (Kb) Conc salt Find mass CH3COONa added to 500ml, 0.10M CH3COOH(acid) pH = 4.5, Ka = 1.8 x 10-5, pKa = 4.74 Conc CH3COO- = 0.0578M → x RMM (82) → 4.74g in 1000ml 2.37g in 500ml 9 2nd method (Ka)1st method (formula) Click here addition base to buffer Click here addition acid to buffer 45.955.414 55.4 ]32.0[ ]50.0[ lg74.4 ][ ][ lg pH pOH pOH salt base pKpOH b 45.955.414 55.4 )1081.2lg( )lg( 5 pH pOH pOH OHpOH 5 5 3 4 423 1081.2 50.0 ))(32.0( 108.1 )( ))(( OH OH NH OHNH K OHNHOHNH b 45.955.414 55.4 ]32.0[ ]50.0[ lg74.4 ][ ][ lg pH pOH pOH salt base pKpOH b 45.955.414 55.4 )1081.2lg( )lg( 5 pH pOH pOH OHpOH 5 5 3 4 423 1081.2 50.0 ))(32.0( 108.1 )( ))(( OH OH NH OHNH K OHNHOHNH b 0578.0][ 24.0 ][ ]10.0[ lg ][ ]10.0[ lg74.45.4 ][ ][ lg 3 3 3 3 COOCH COOCH COOCH COOCH acid pKpH a 5.4 10 )lg(5.4 )lg( H H HpH MCOOCH COOCH COOHCH HCOOCH K HCOOCHCOOHCH a 0578.0][ )10.0( )10)(( 108.1 )( ))(( 3 5.4 35 3 3 33 Conc [H+]
- 23. Given 100ml of 0.05M HCOOH , what vol 0.05M NaOH needed to make pH buffer 4.23. pKa = 3.75 Buffer Calculation 100ml buffer contain 0.10M butanoic acid and 0.20 M sodium butanoate. What pH change when 2.0cm3, 0.10M HCI added. pKa = 4.82 10 11 After adding acid Click here addition base to buffer Click here addition acid to buffer 3 75 ][ ][ lg75.323.4 ][ ][ lg cmvol salt acid salt acid pKpH a 1000 05.0 .... v saltmolNaOHmol NaOH + HCOOH → HCOONa + H2O All NaOH react to form salt Mol NaOH react = Mol salt form Mol acid remain = Mol initial – mol react 1000 05.0 1000 10005.0 .. v acidmol 12.5 ]020.0[ ]010.0[ lg82.4 ][ ][ lg pH pH salt acid pKpH a How would adding 100ml DI water affect pH. Before adding acid 0002.0 1000 10.00.2 .... addedacidmol 0102.00002.001.0.. acidmolTotal 0198.00002.002.0.. 0002.0 1000 10.00.2 .... leftsaltmol reactedsaltmol After adding acid 10.5 ]0198.0[ ]0102.0[ lg82.4 ][ ][ lg pH pH salt acid pKpH a Buffer do not change pH on dilution
