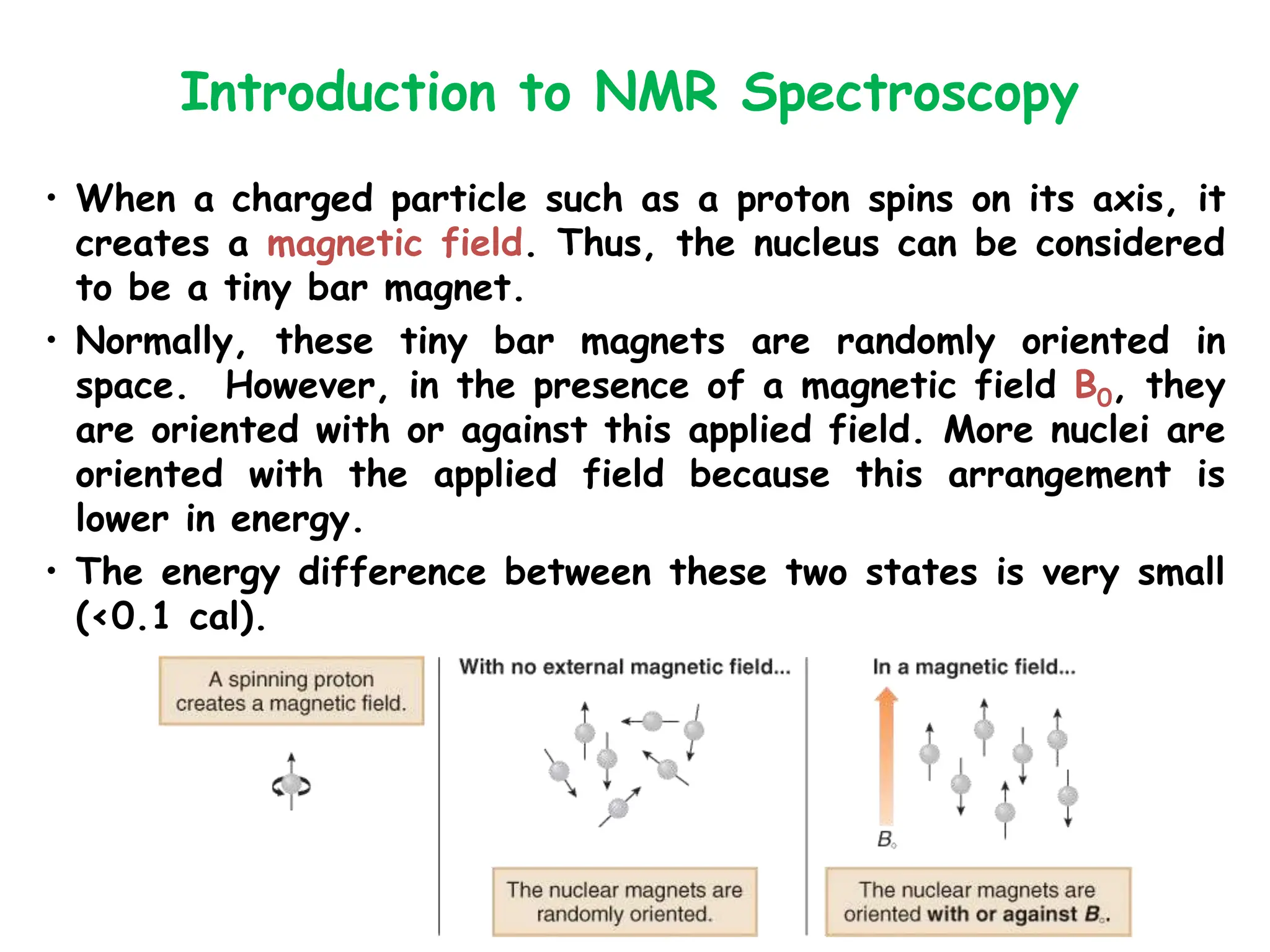
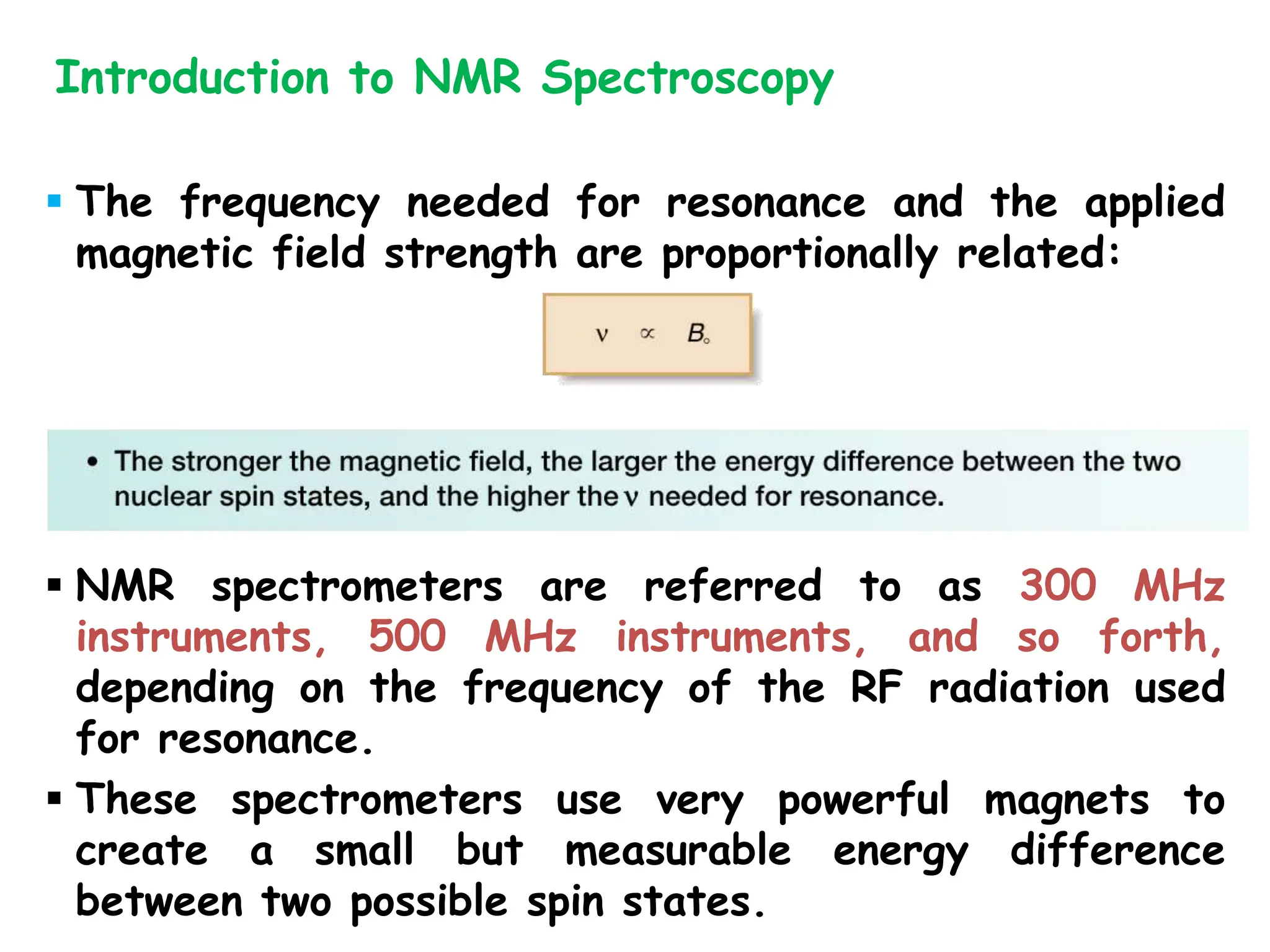
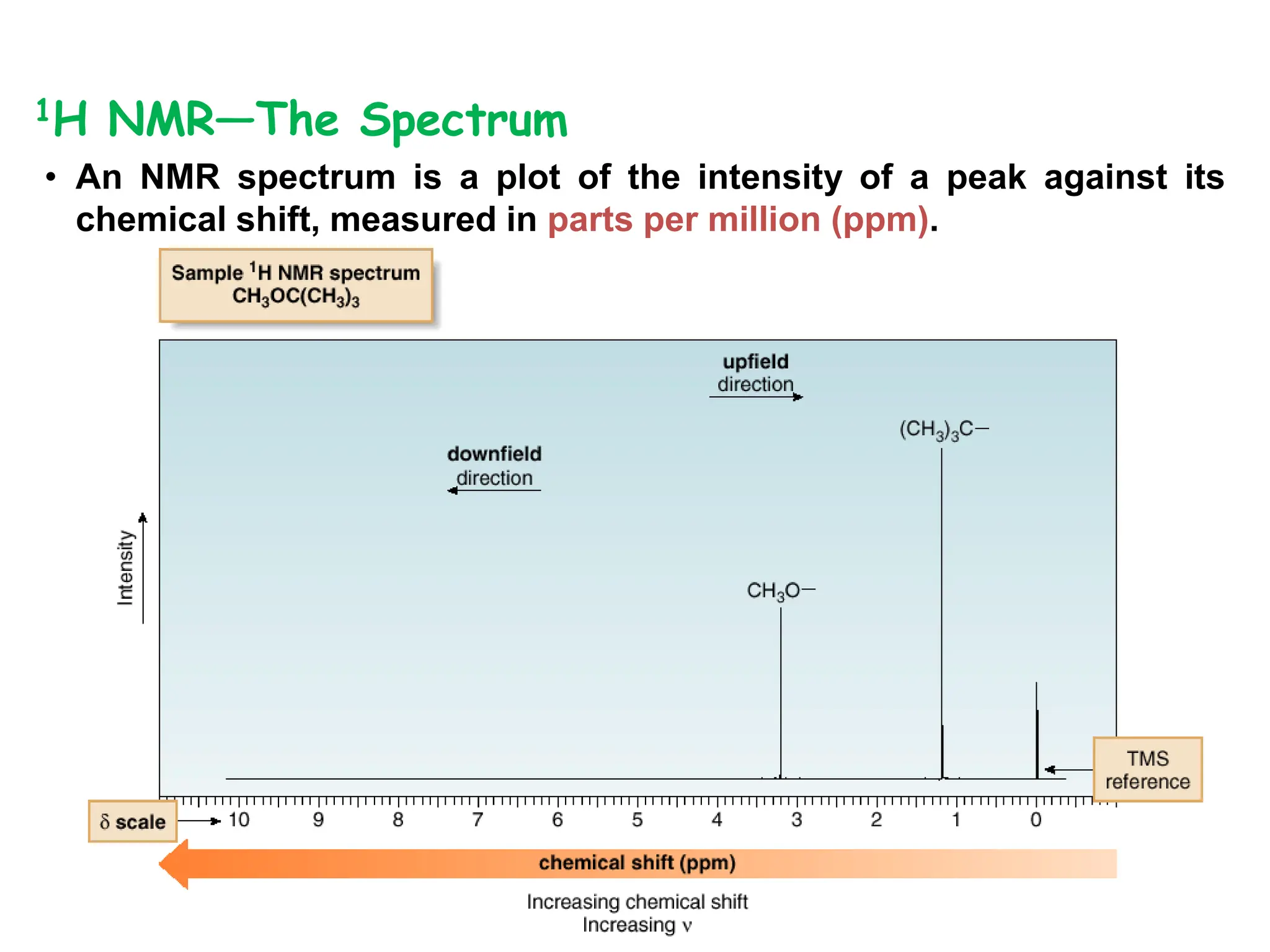
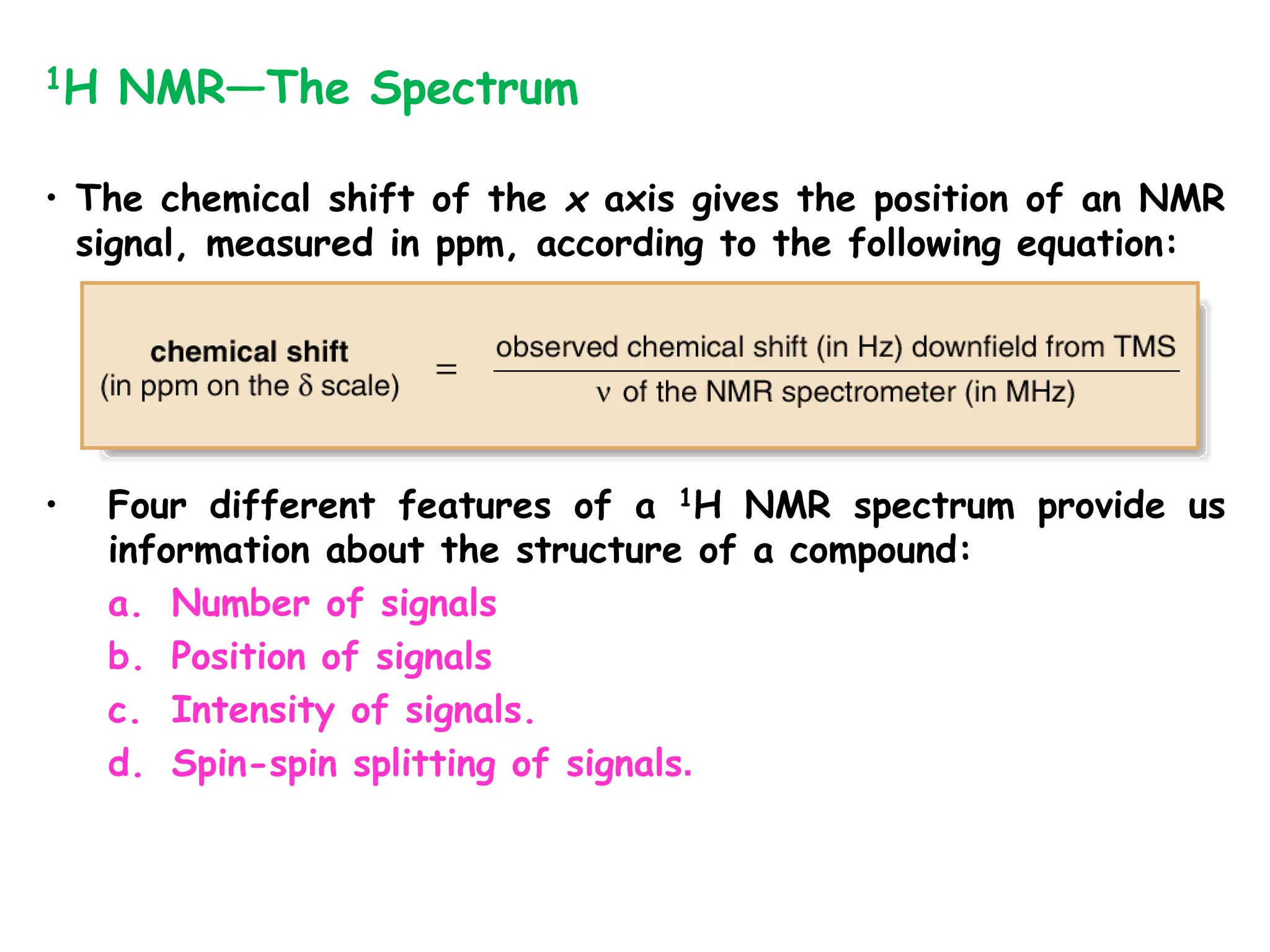
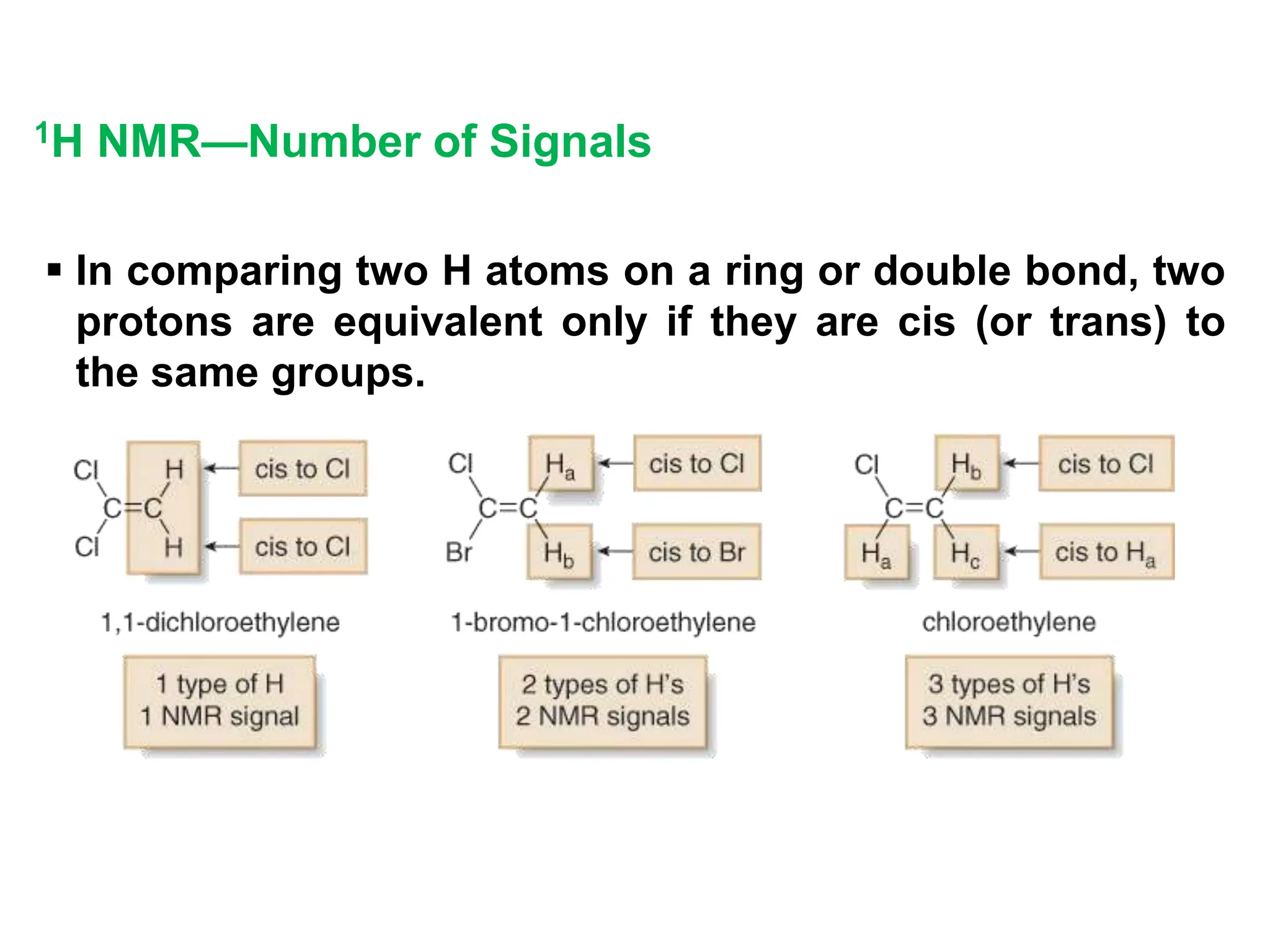
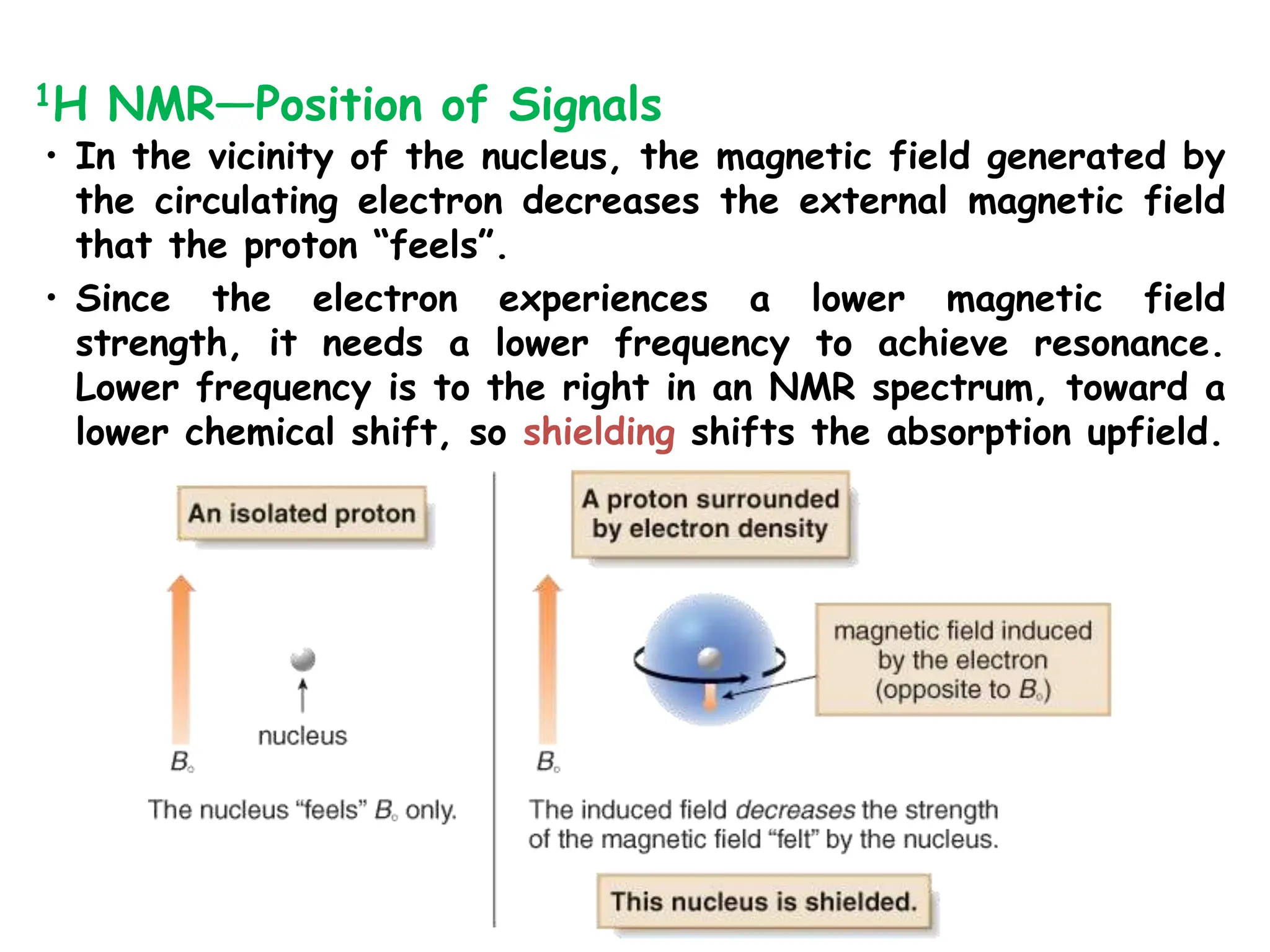
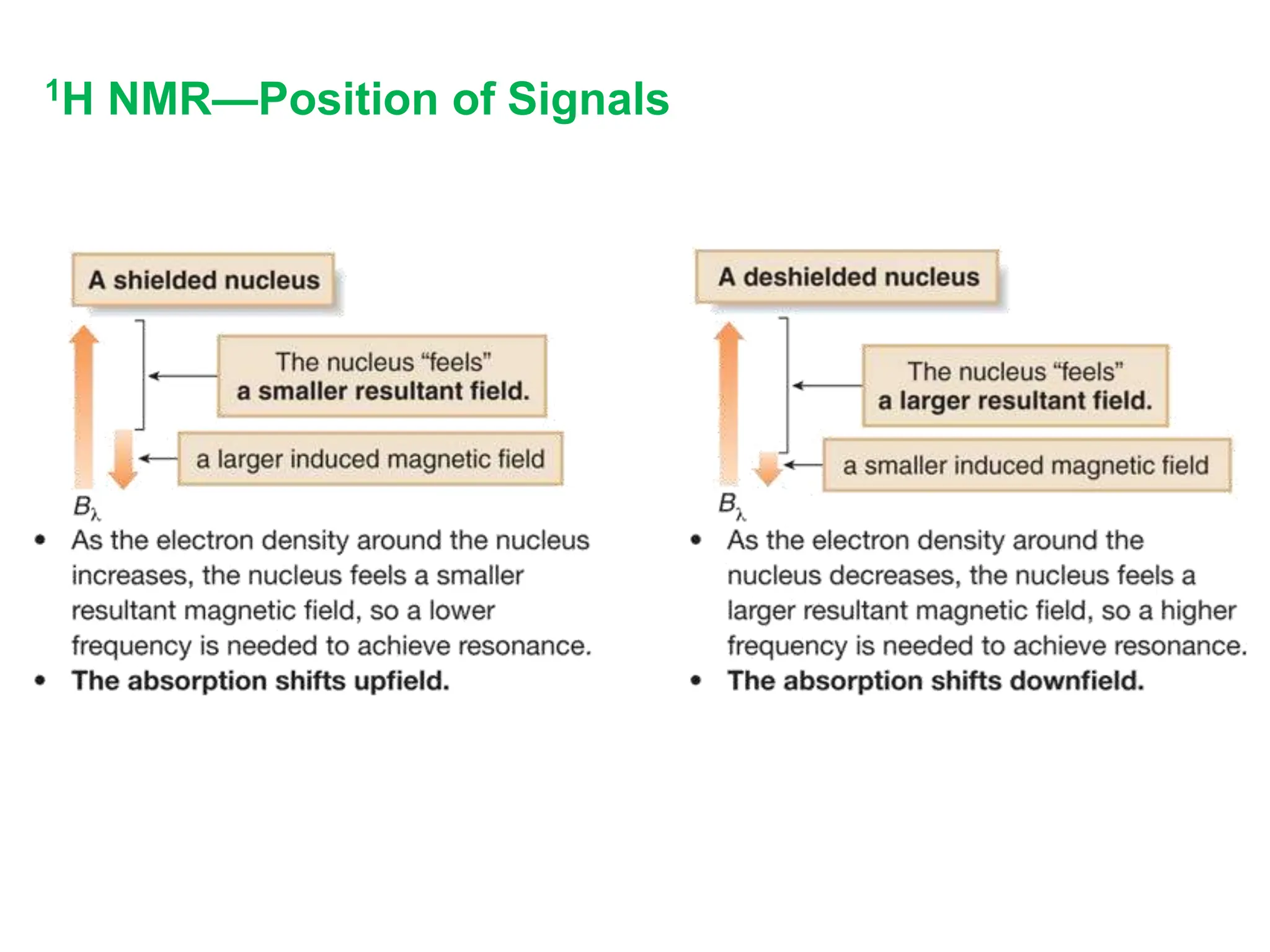
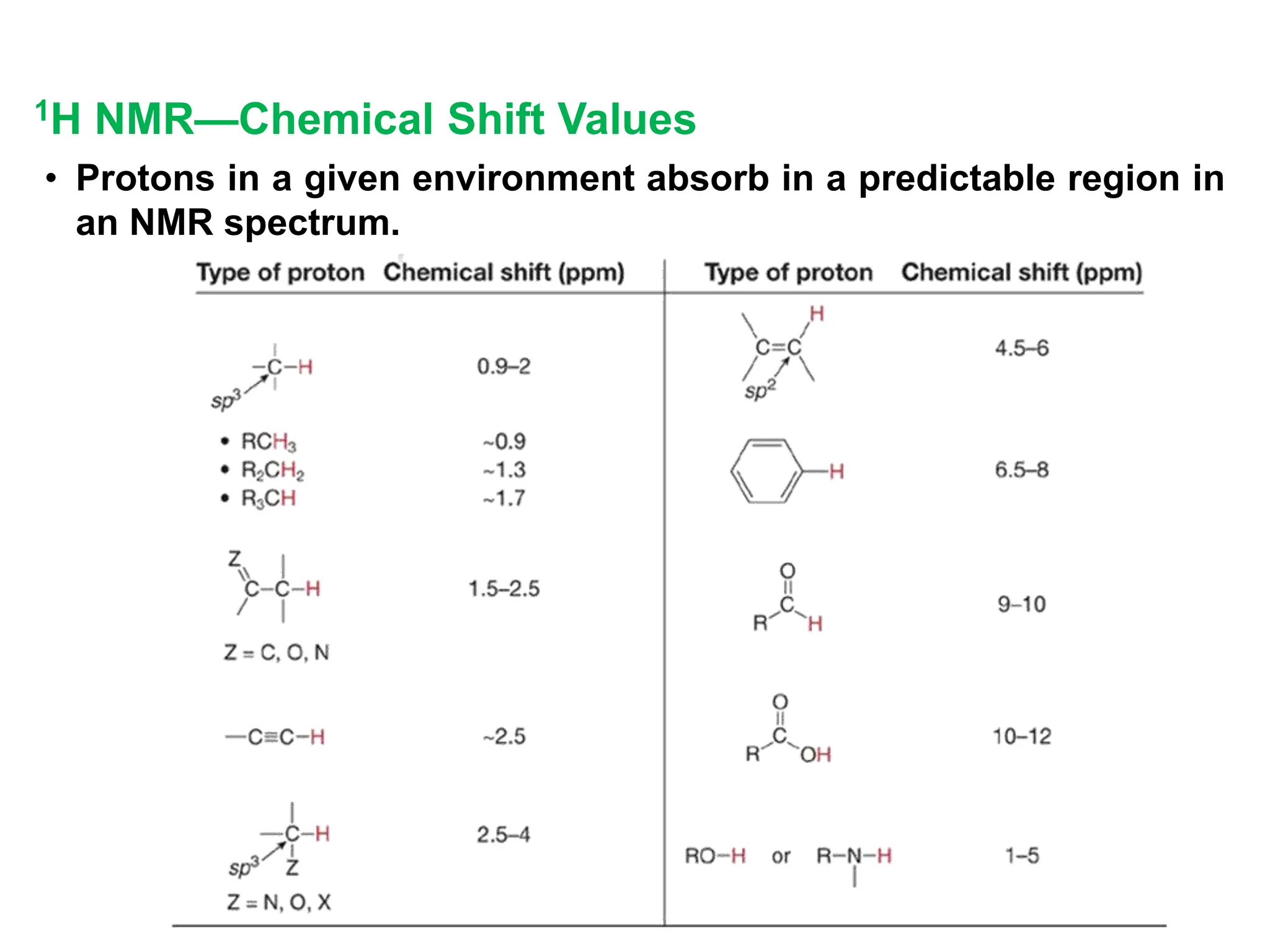
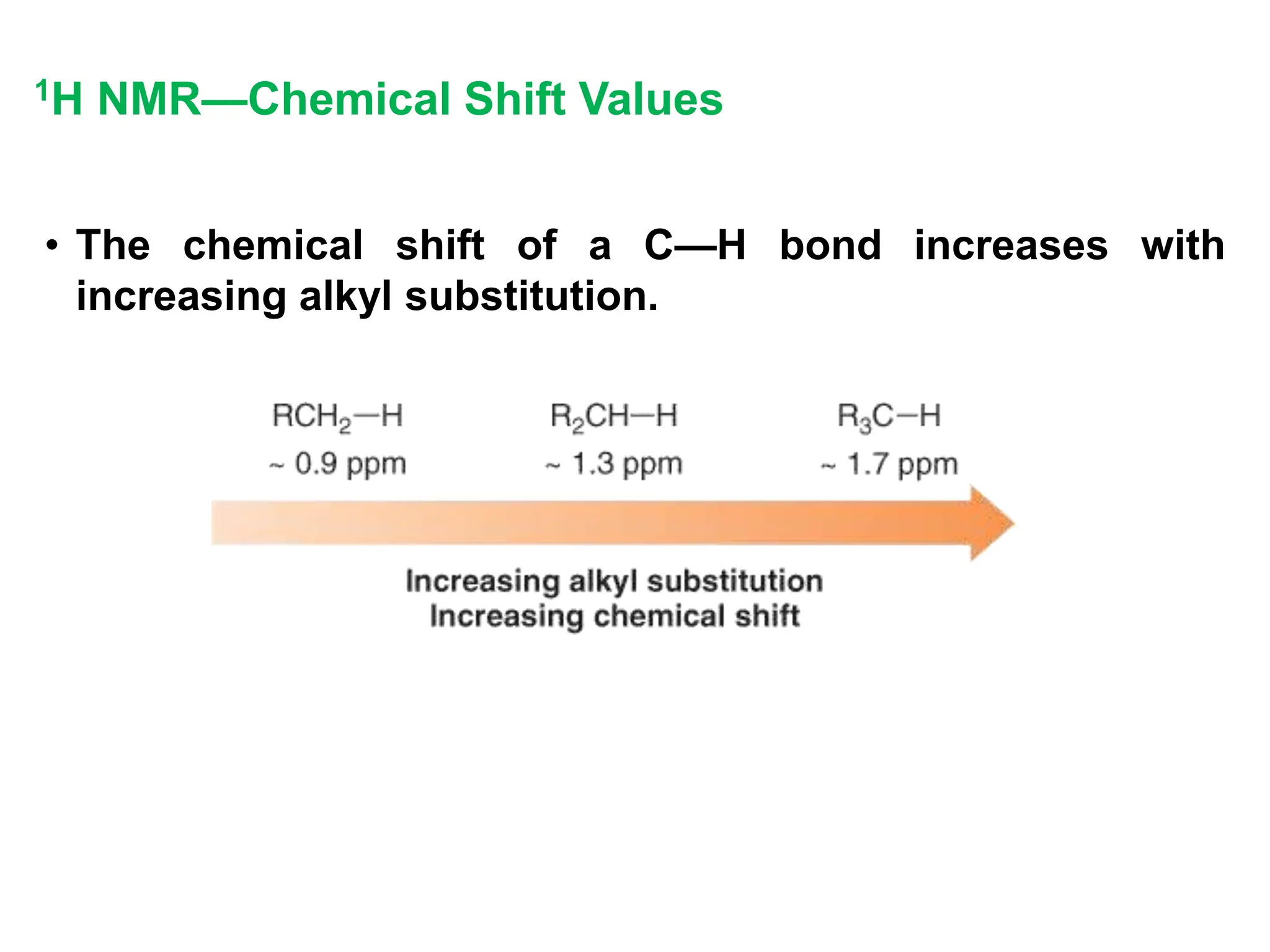
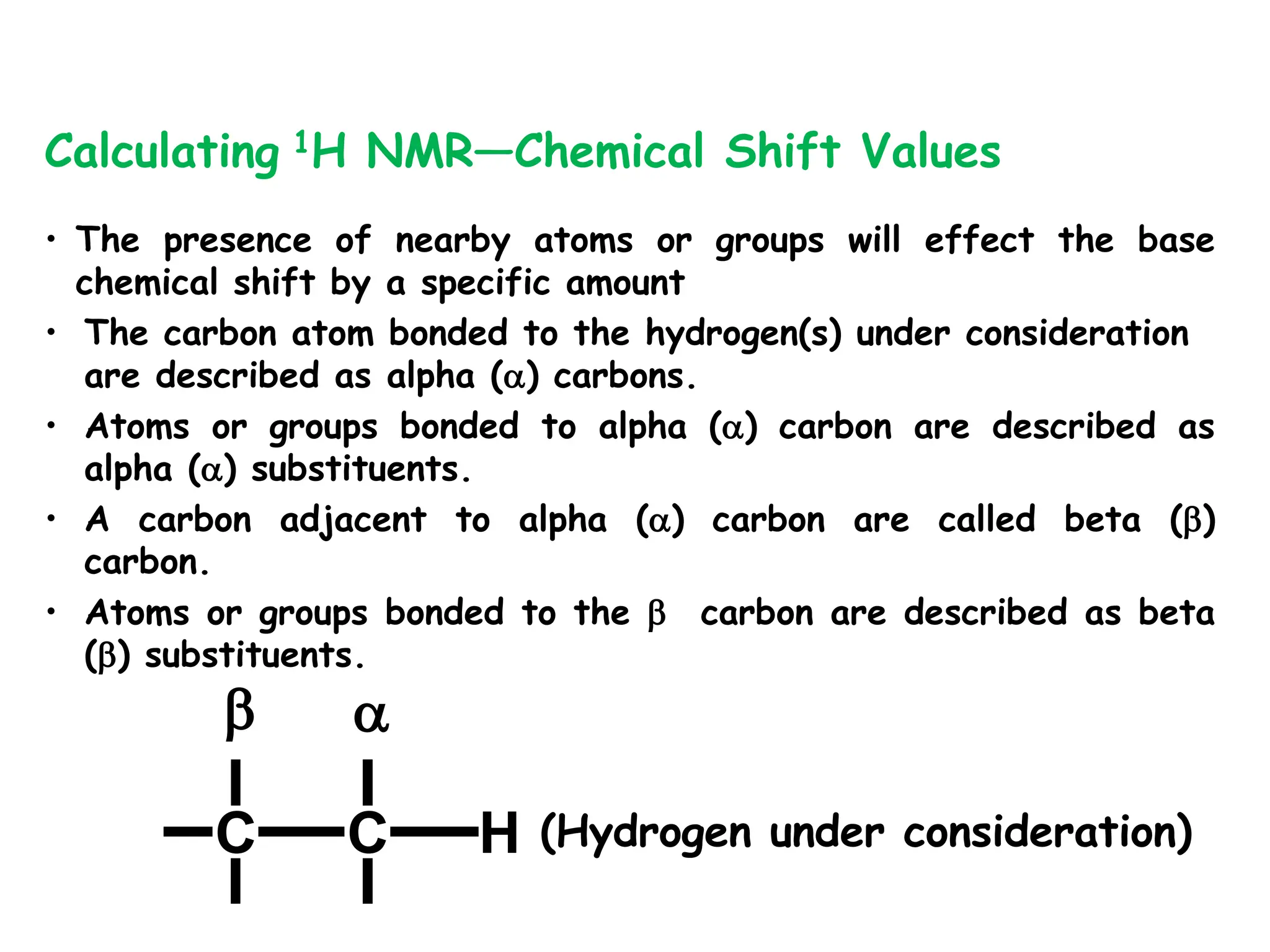
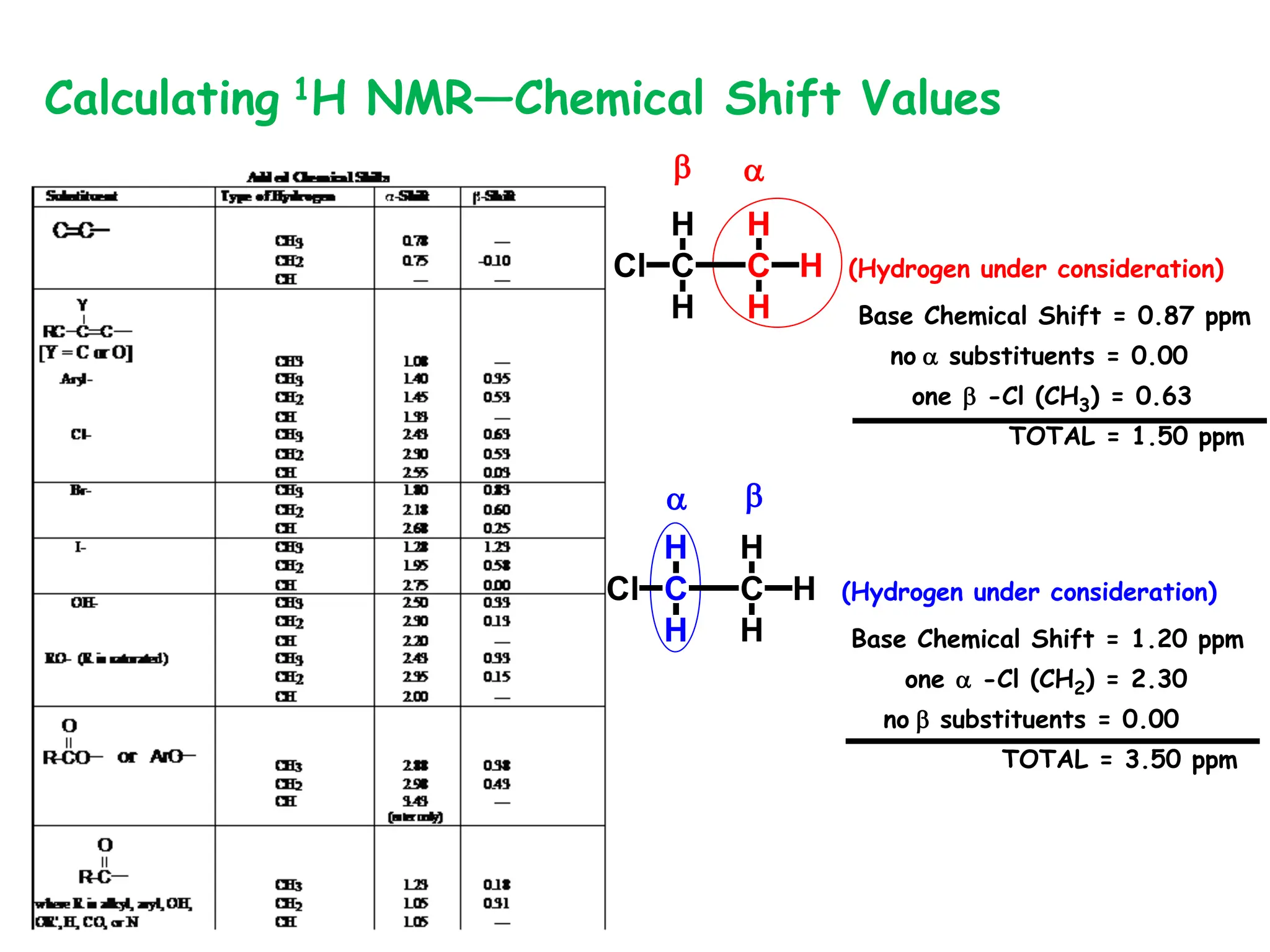
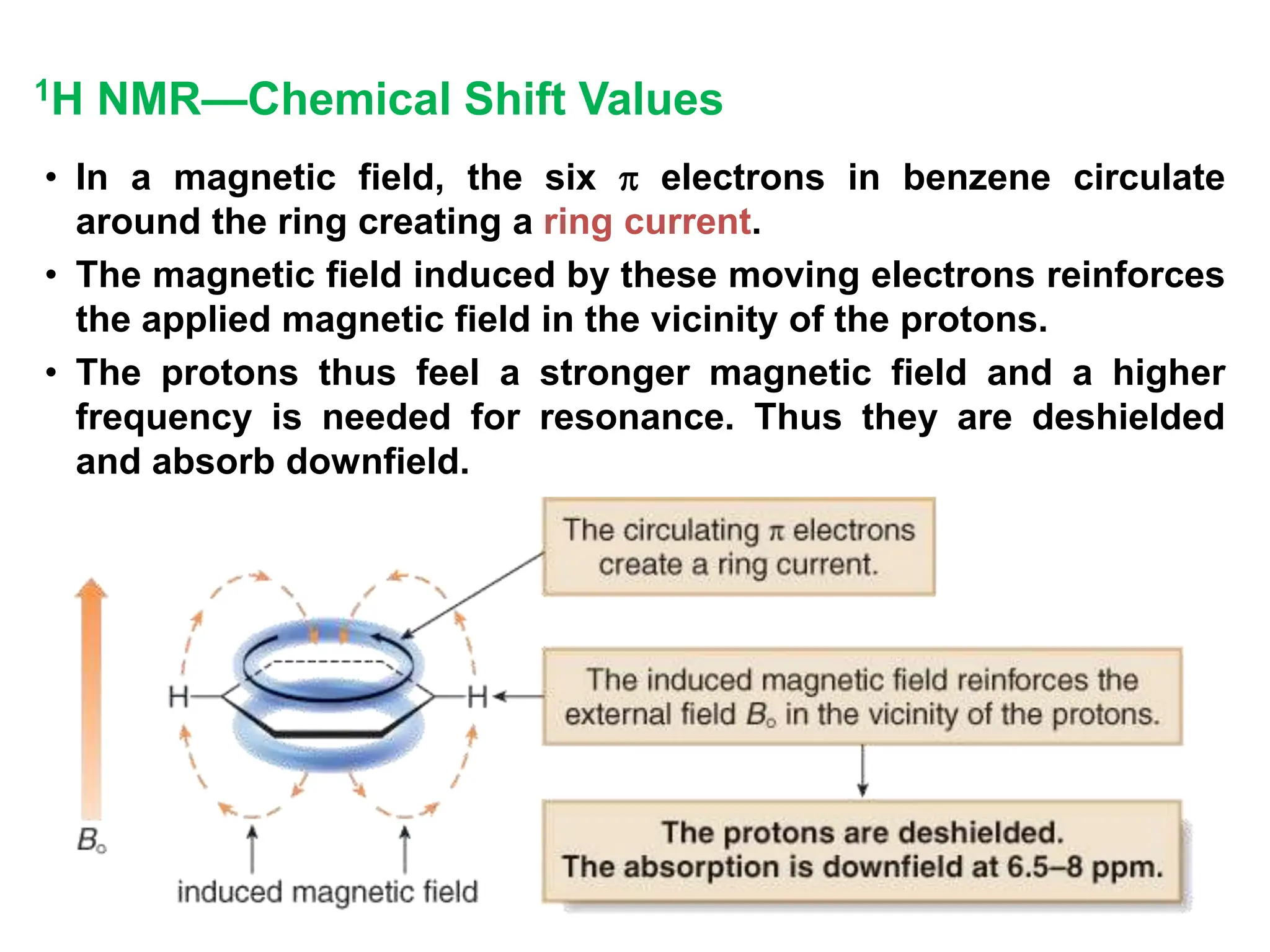
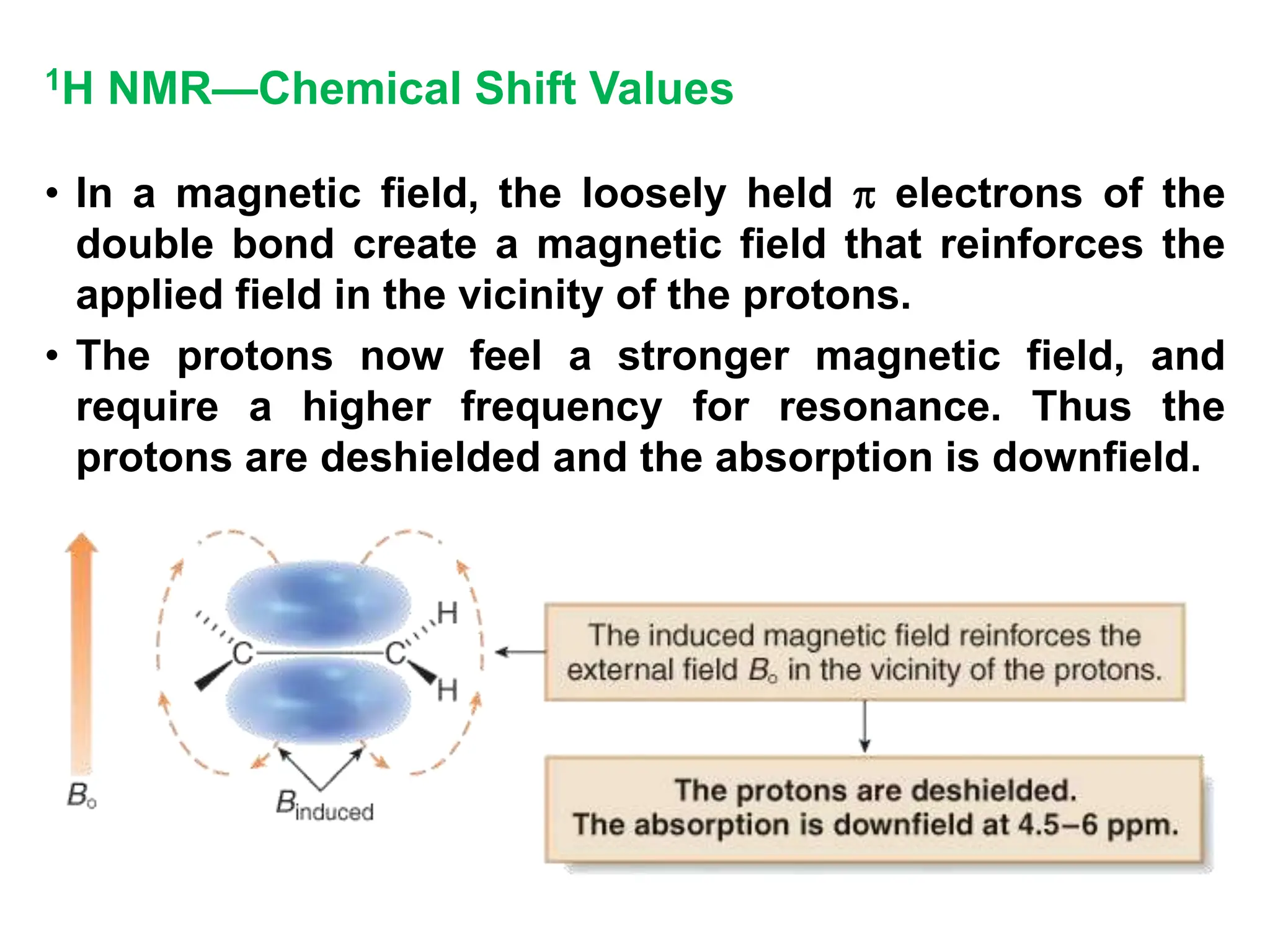
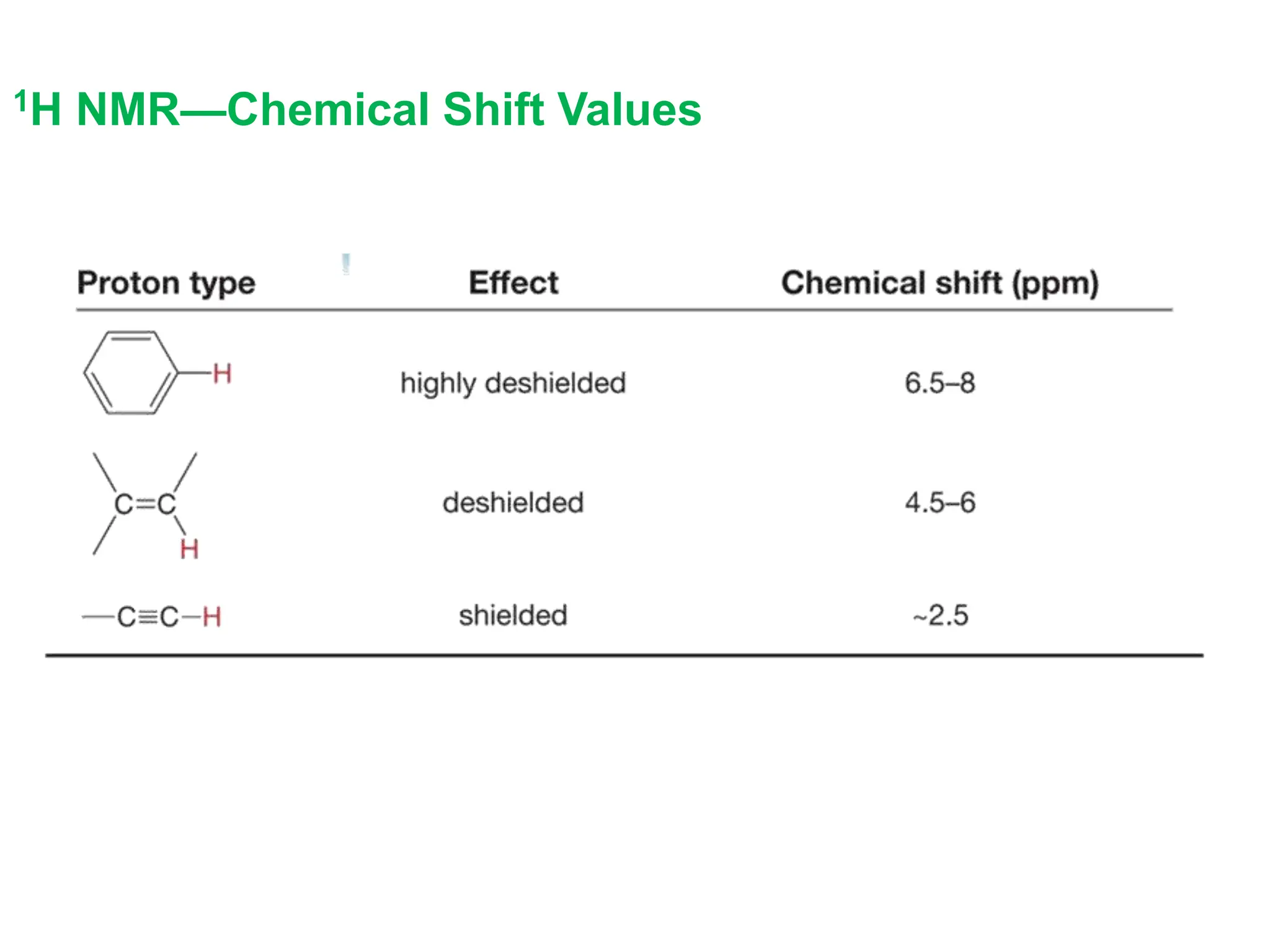
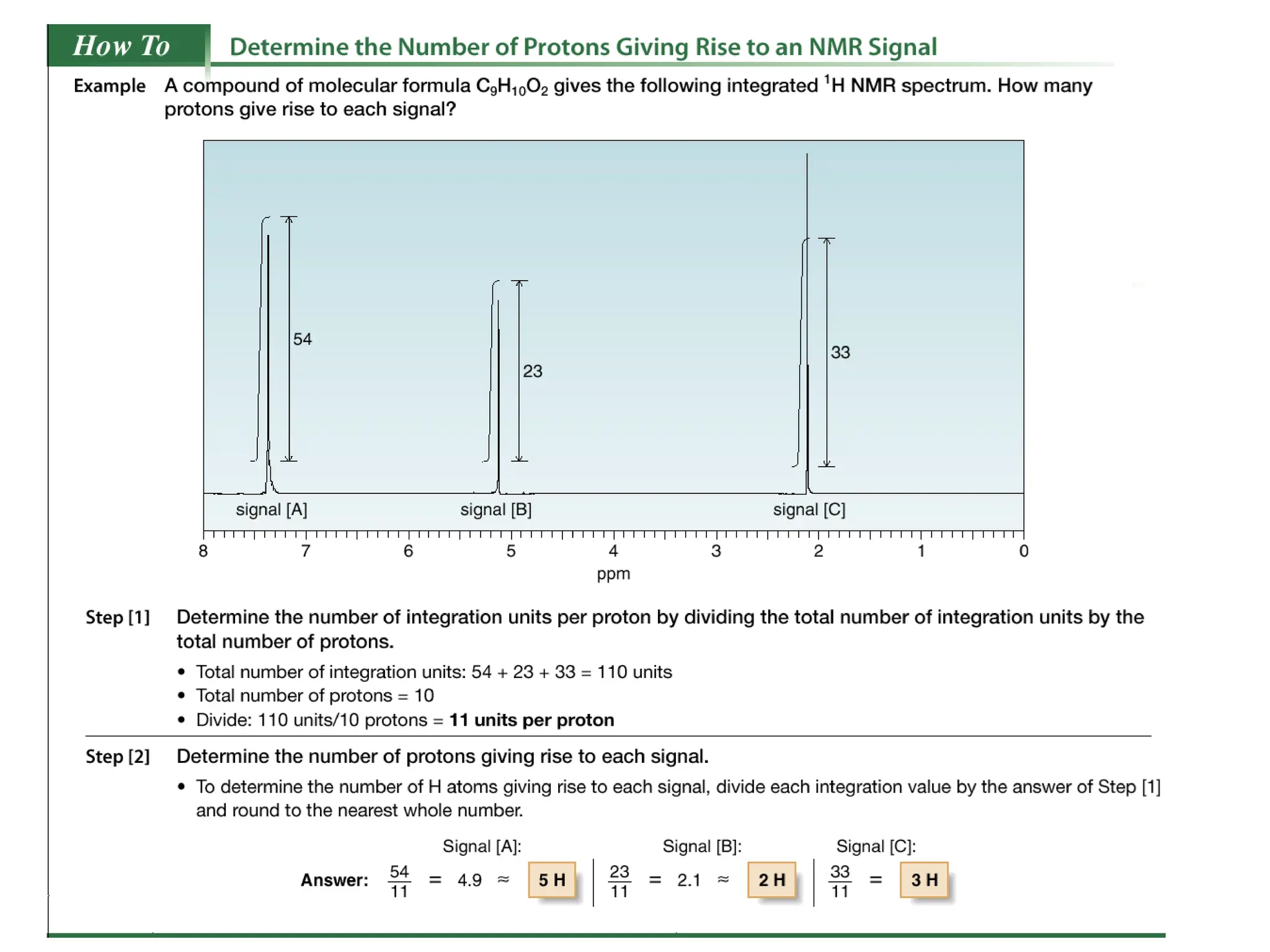
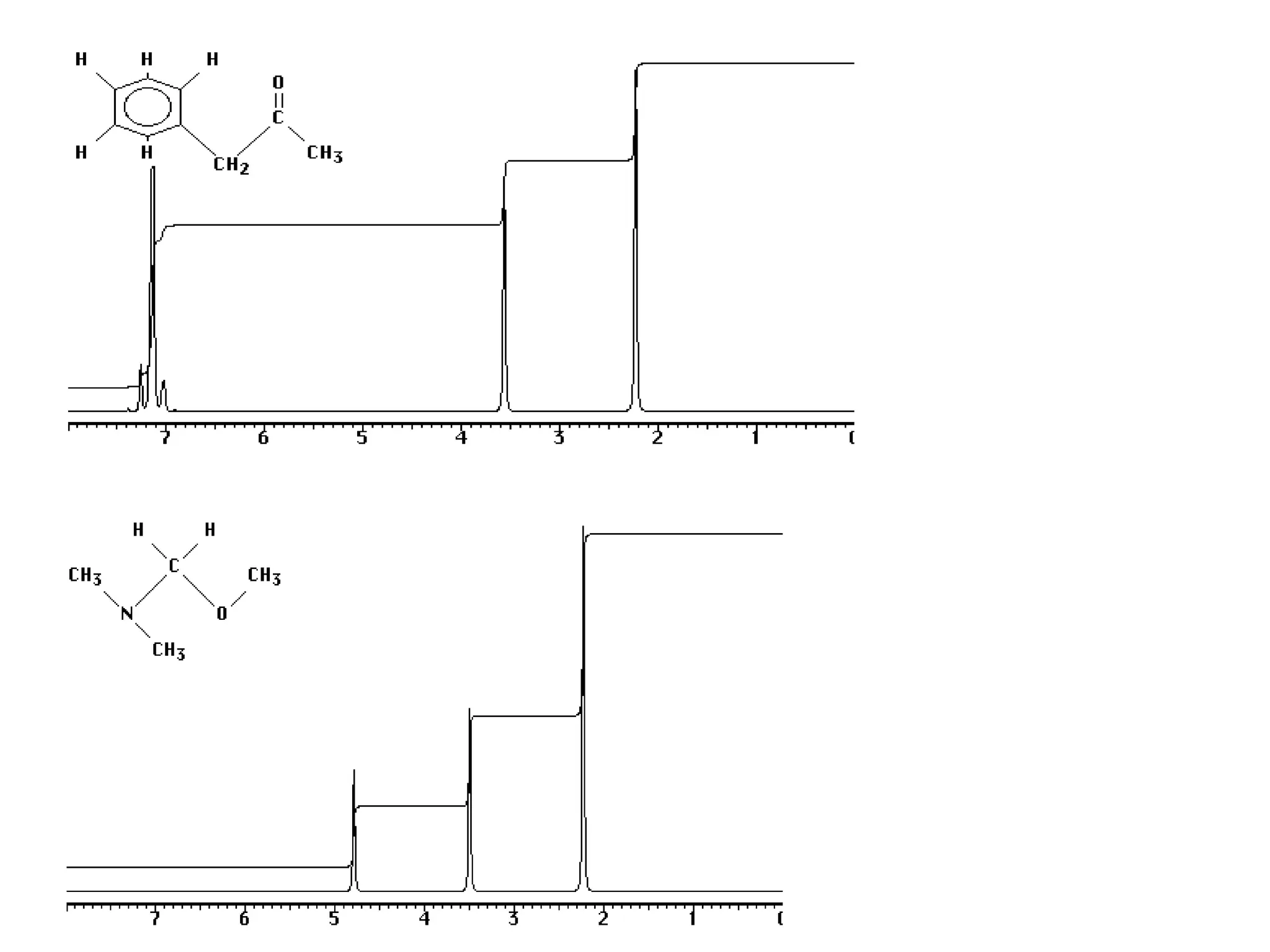
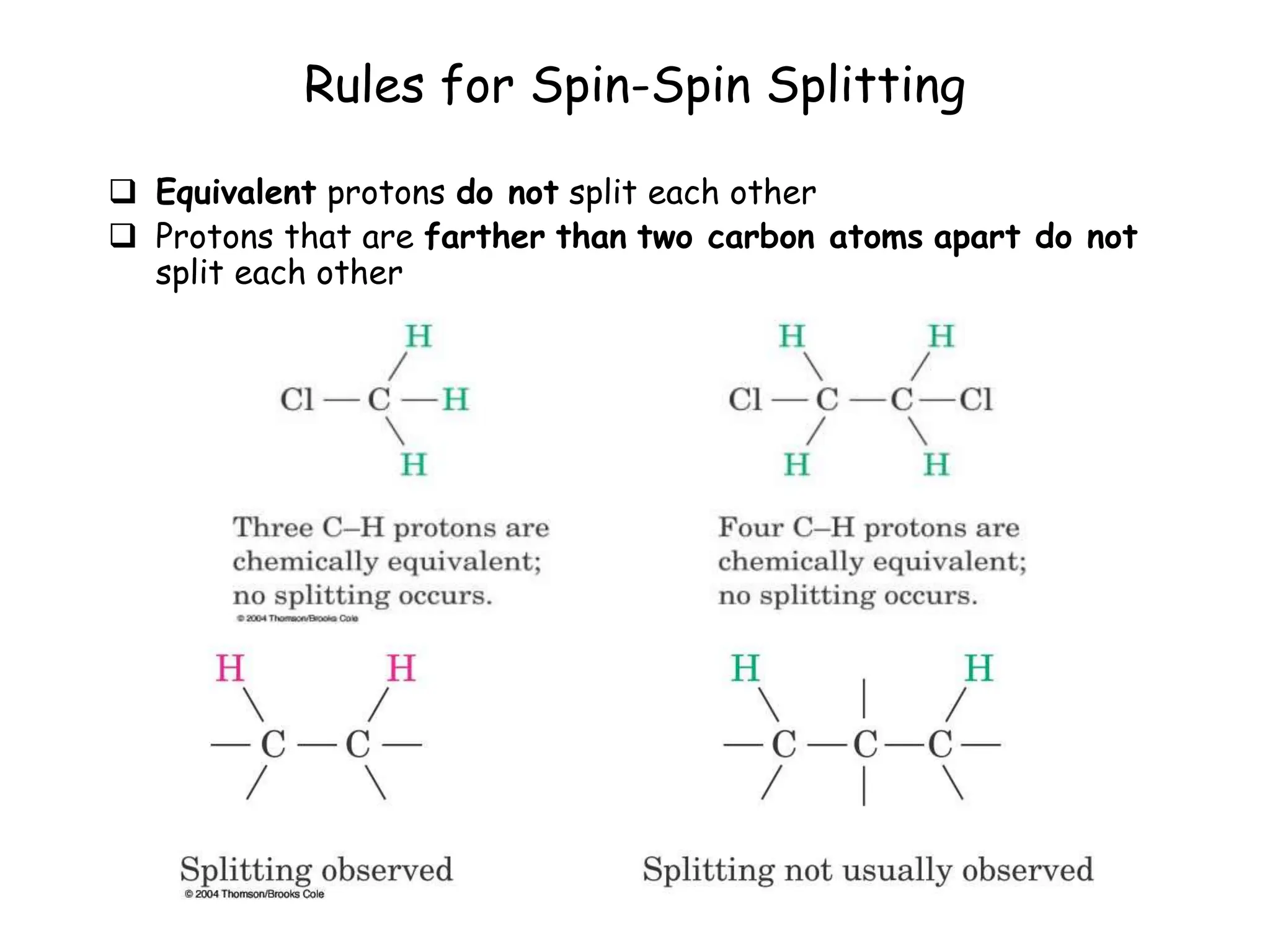
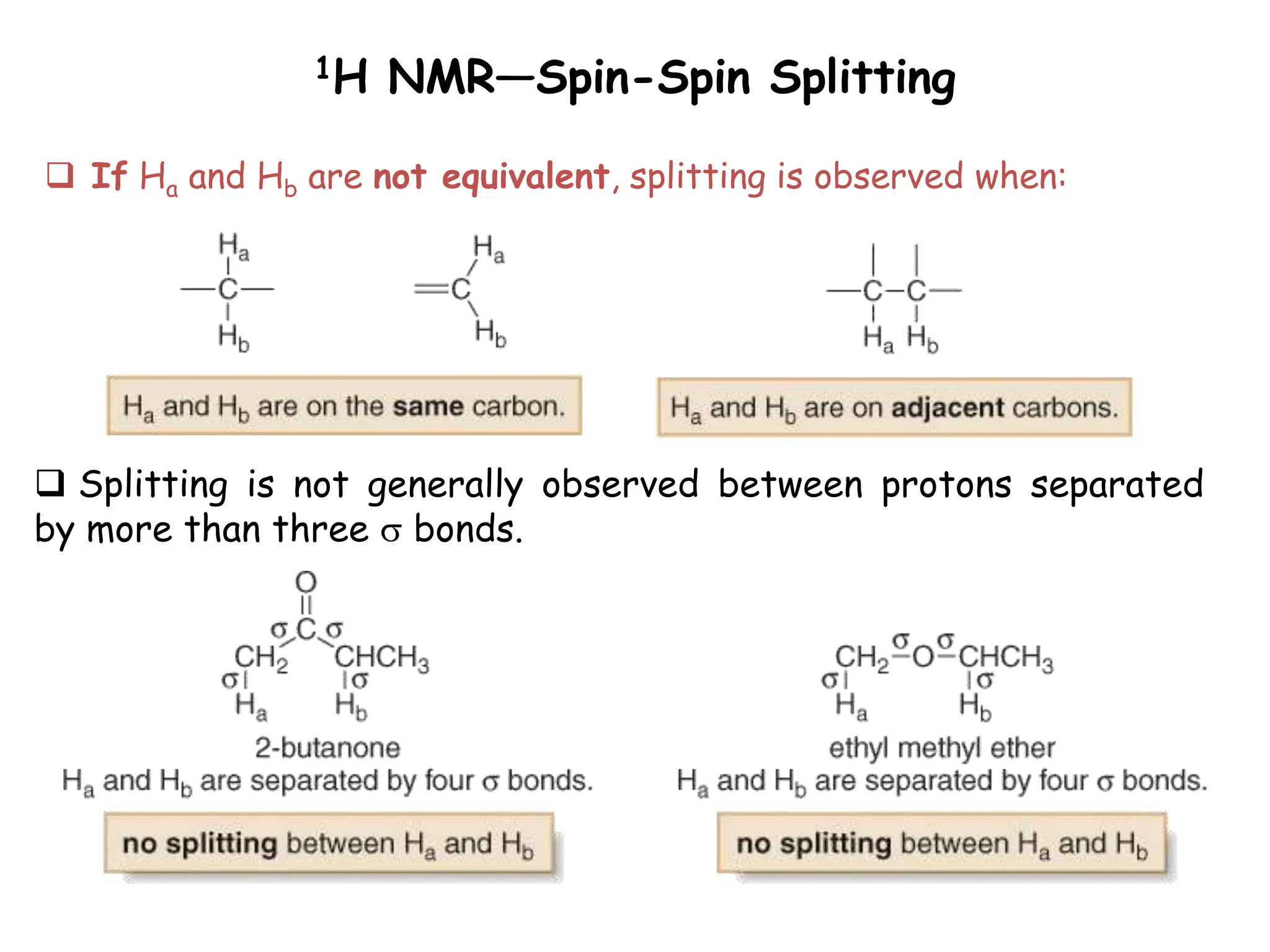
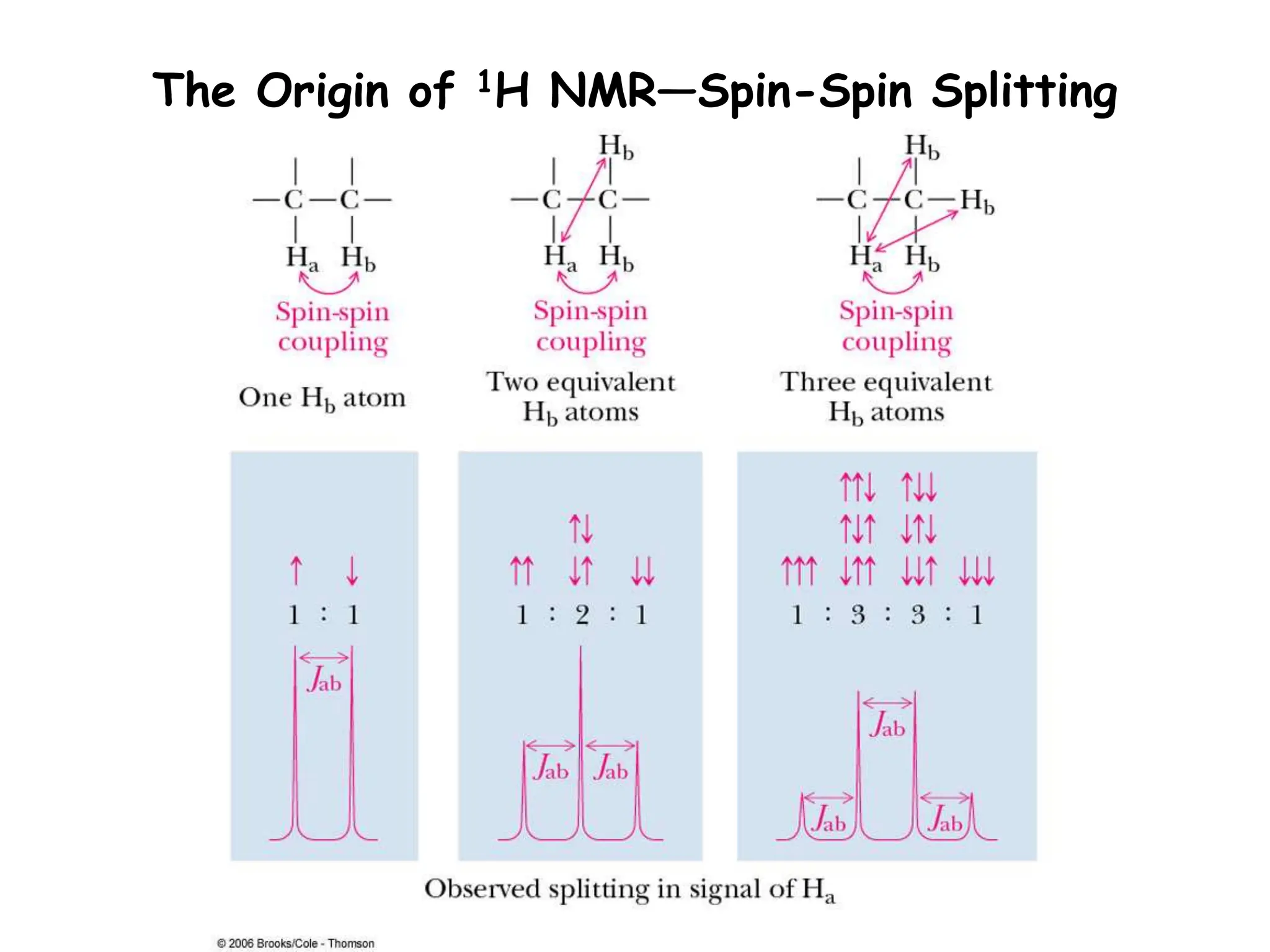
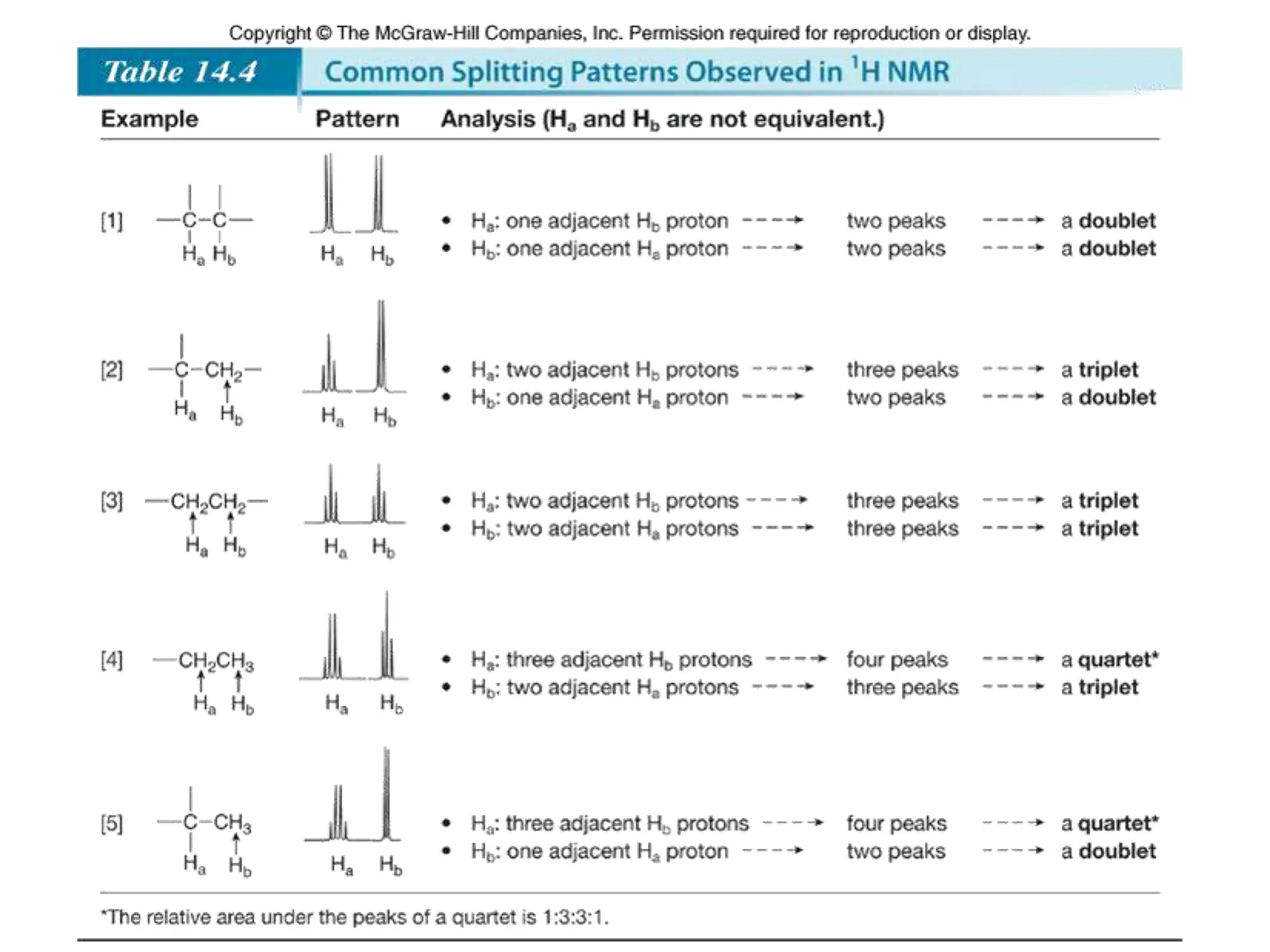
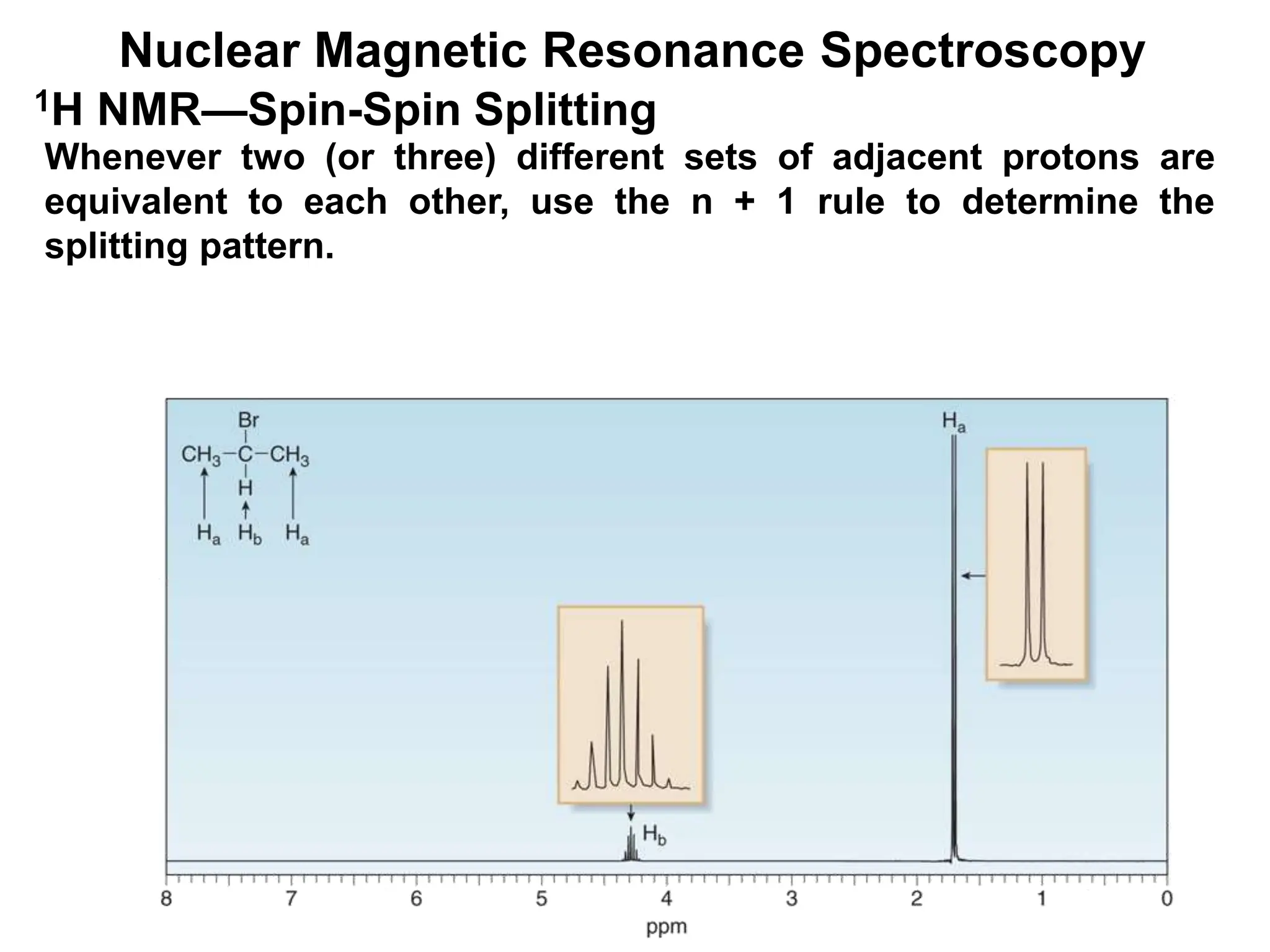
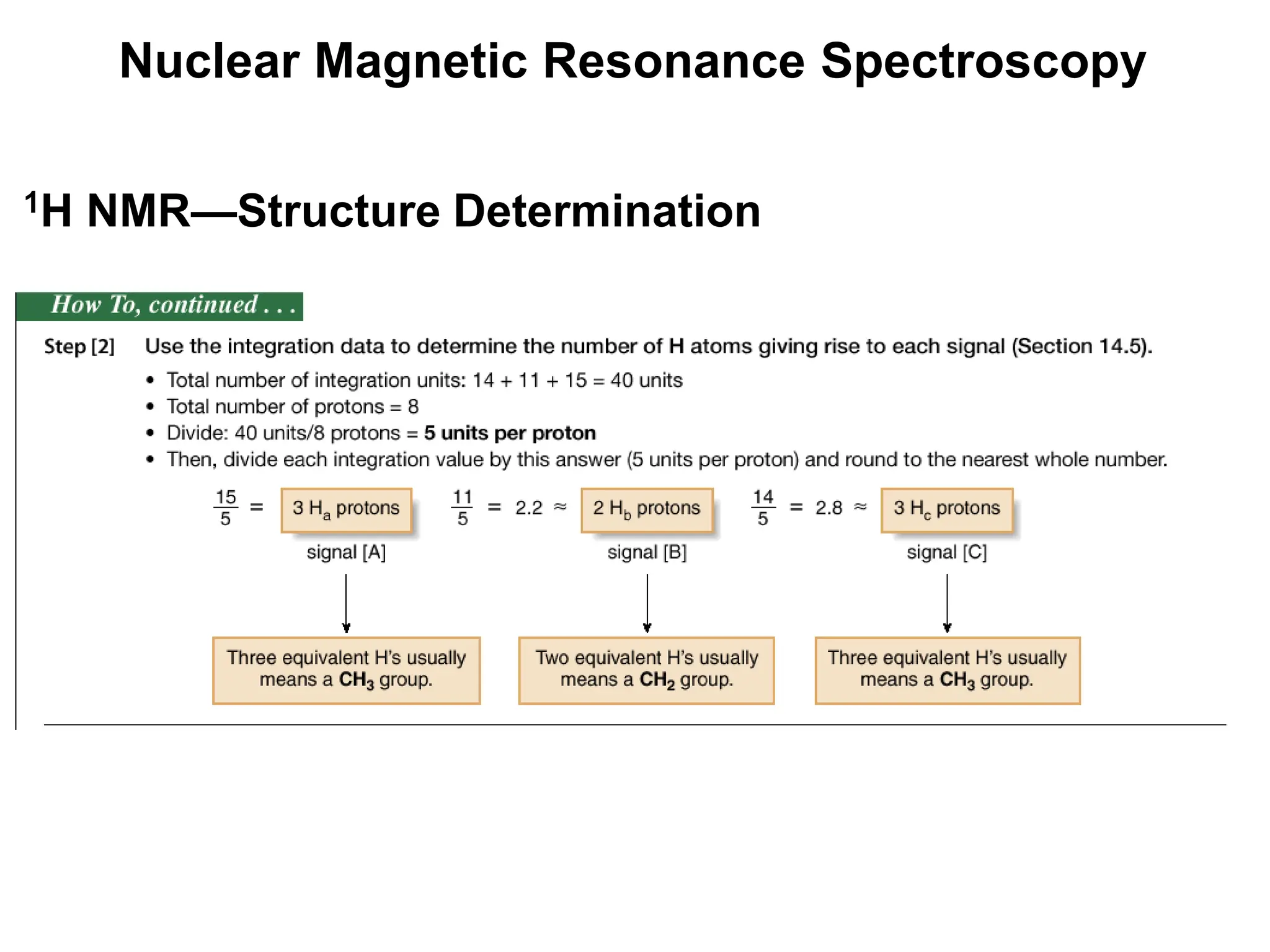
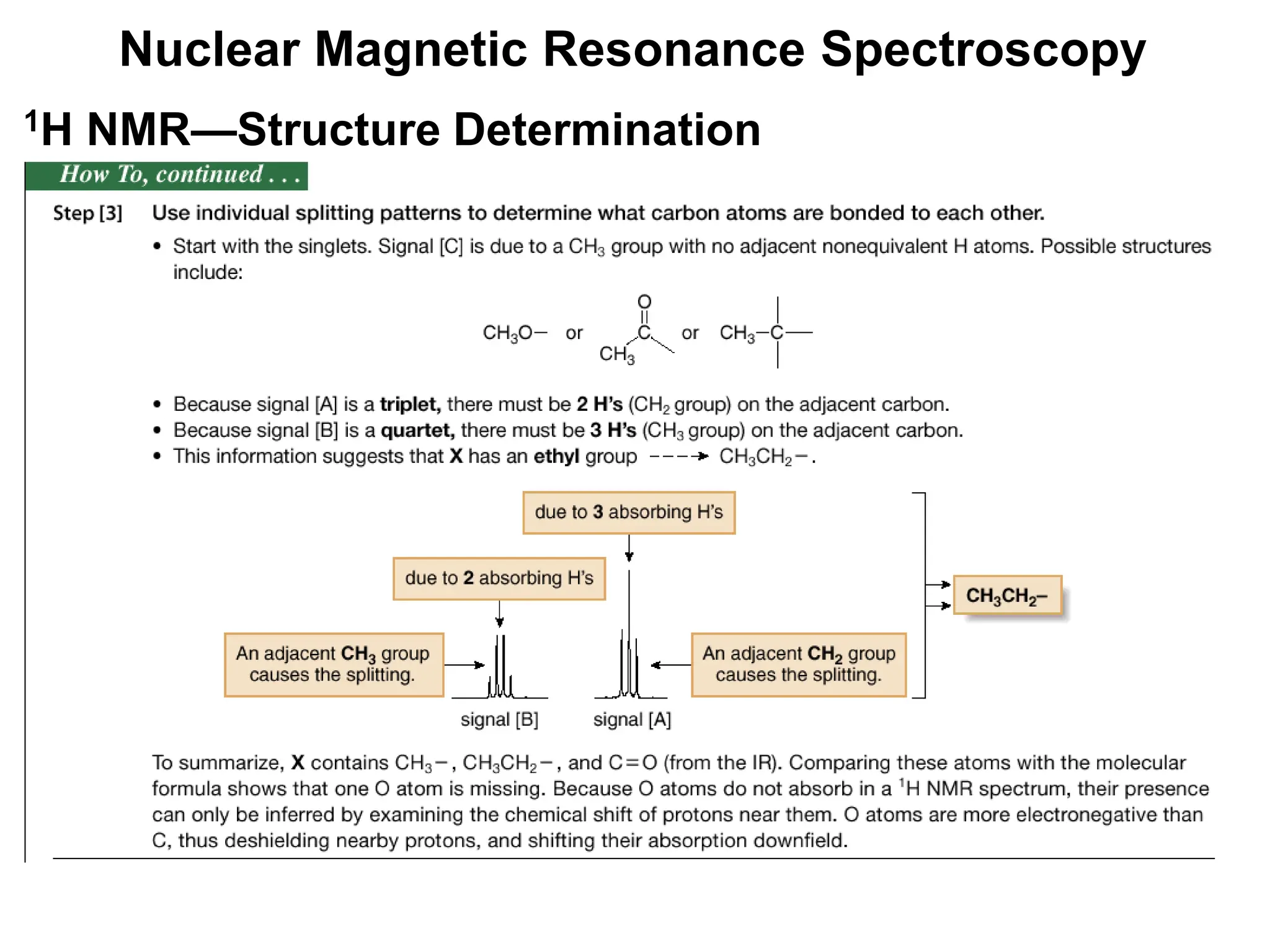
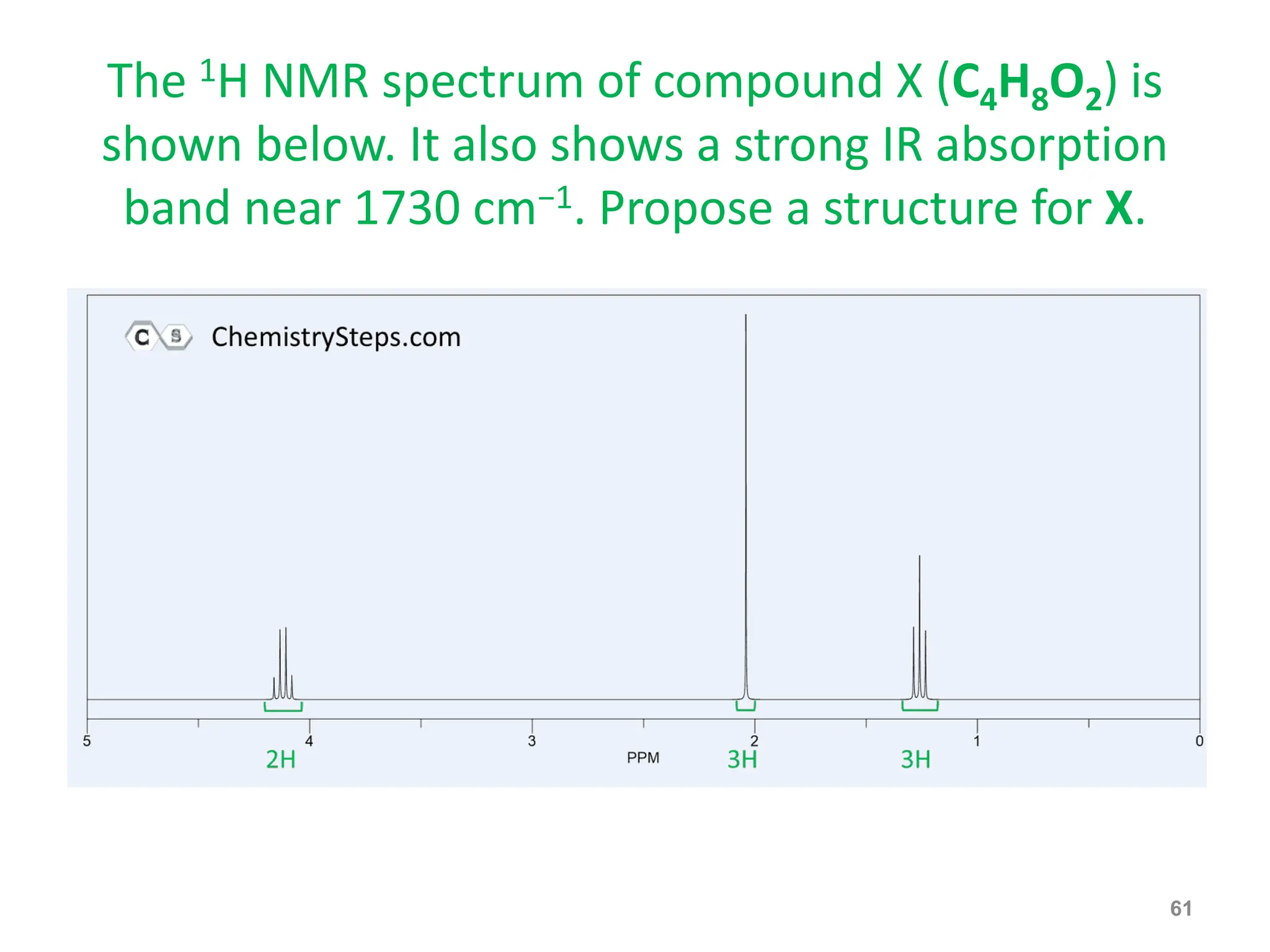
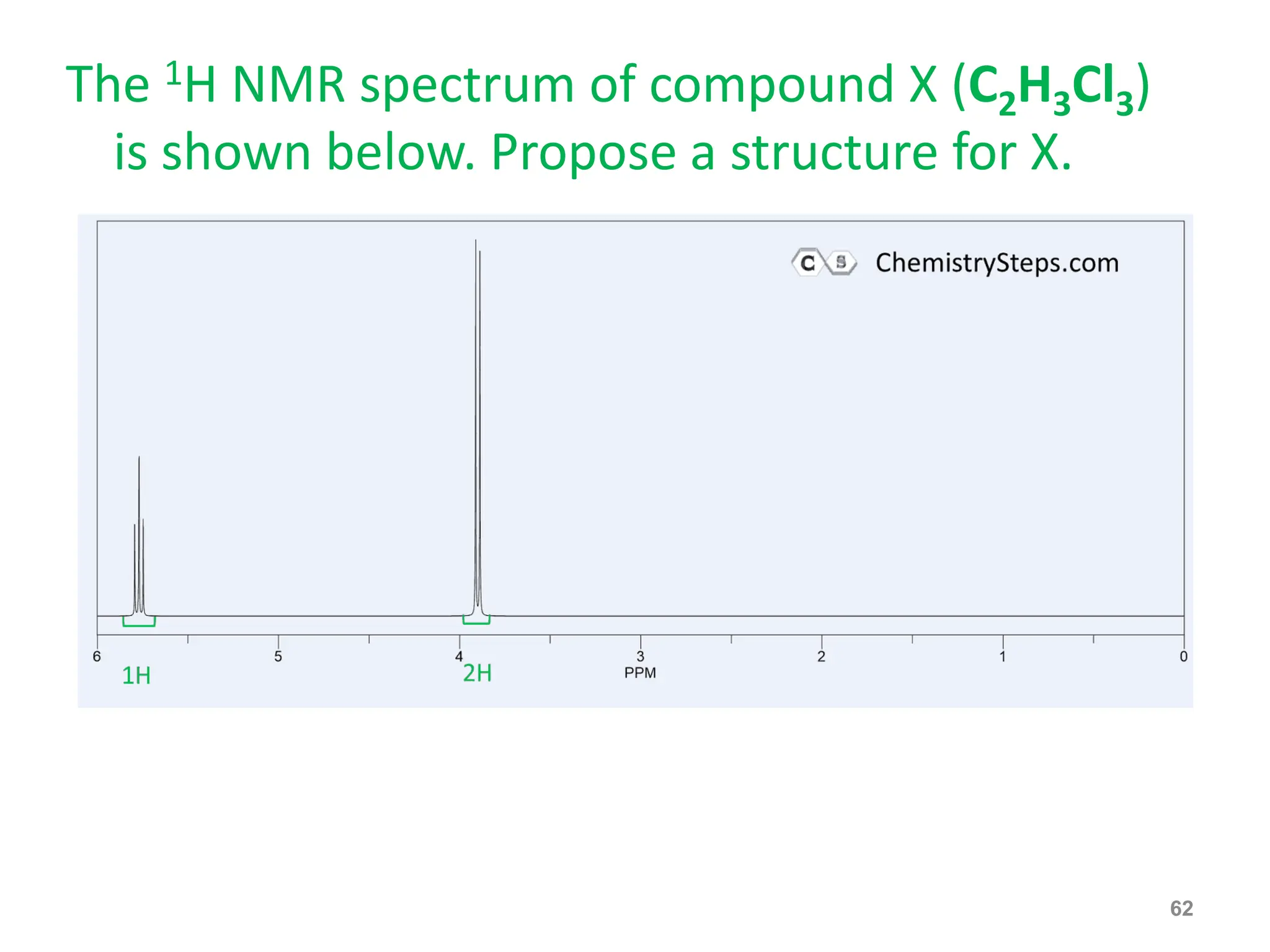
The document provides an introduction to NMR spectroscopy. It discusses how 1H and 13C NMR can be used to characterize organic molecules by identifying carbon-hydrogen frameworks. It explains the basic principles of NMR including how radio waves interact with nuclei to cause spin flipping between energy states. It also discusses how NMR spectra are obtained and analyzed, focusing on 1H NMR. Key points covered include the number, position, intensity and splitting of 1H NMR signals and how they provide structural information.