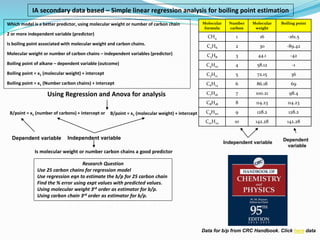
IA data based, boiling point estimation using molecular weight and carbon chain.
- 1. Which model is a better predictor, using molecular weight or number of carbon chain 2 or more independent variable (predictor) Is boiling point associated with molecular weight and carbon chains. Molecular weight or number of carbon chains – independent variables (predictor) Boiling point of alkane – dependent variable (outcome) Boiling point = x1 (molecular weight) + intercept Boiling point = x1 (Number carbon chains) + intercept Using Regression and Anova for analysis Independent variable Dependent variable Is molecular weight or number carbon chains a good predictor Independent variable Dependent variable Data for b/p from CRC Handbook. Click here data IA secondary data based – Simple linear regression analysis for boiling point estimation B/point = x1 (molecular weight) + intercept B/point = x1 (number of carbons) + intercept or Research Question Use 25 carbon chains for regression model Use regression eqn to estimate the b/p for 25 carbon chain Find the % error using expt values with predicted values. Using molecular weight 3rd order as estimator for b/p. Using carbon chain 3rd order as estimator for b/p. Molecular formula Number carbon Molecular weight Boiling point CH4 1 16 -161.5 C2H6 2 30 -89.42 C3H8 3 44.1 -42 C4H10 4 58.12 -1 C5H12 5 72.15 36 C6H14 6 86.18 69 C7H16 7 100.21 98.4 C8H18 8 114.23 114.23 C9H20 9 128.2 128.2 C10H22 10 142.28 142.28
- 2. • member differ by CH2 gp • same functional group • similar chemical properties • chemical formula CnH2n+2 • end with ane Class Functional gp Suffix Example Formula Alkane C - C - ane ethane CnH2n+2 Homologous Series Class Functional Suffix Example Formula Alkene Alkenyl - ene ethene CnH2n H H ׀ ׀ H - C – C – H ׀ ׀ H H • member differ by CH2 gp • same functional group • similar chemical properties • chemical formula CnH2n • end with ene H ׀ H - C – H ׀ H H H H ׀ ׀ ׀ H - C – C – C – H ׀ ׀ ׀ H H H H H H H ׀ ׀ ׀ ׀ H - C – C – C – C – H ׀ ׀ ׀ ׀ H H H H Number carbon Word IUPAC name Structure formula Molecular formula 1 Meth Methane CH4 CH4 2 Eth Ethane CH3CH3 C2H6 3 Prop Propane CH3CH2CH3 C3H8 4 But Butane CH3(CH2)2CH3 C4H10 5 Pent Pentane CH3(CH2)3CH3 C5H12 6 Hex Hexane CH3(CH2)4CH3 C6H14 7 Hept Heptane CH3(CH2)5CH3 C7H16 8 Oct Octane CH3(CH2)6CH3 C8H18 9 Non Nonane CH3(CH2)7CH3 C9H20 10 Dec Decane CH3(CH2)8CH3 C10H22 methane ethane propane butane Saturated hydrocarbon (C – C single bond) Number carbon IUPAC name Structure formula Molecular formula 2 Ethene CH2CH2 C2H4 3 Propene CH2=CHCH3 C3H6 4 Butene CH2=CHCH2CH3 C4H8 5 Pentene CH2=CH(CH2)2CH3 C5H10 6 Hexene CH2=CH(CH2)3CH3 C6H12 7 Heptene CH2=CH(CH2)4CH3 C7H14 8 Octene CH2=CH(CH2)5CH3 C8H16 9 Nonene CH2=CH(CH2)6CH3 C9H18 10 Decene CH2=CH(CH2)7CH3 C10H20 H H ׀ ׀ C = C ׀ ׀ H H H H H ׀ ׀ ׀ C = C – C - H ׀ ׀ H H H H H H ׀ ׀ ׀ ׀ C = C – C – C - H ׀ ׀ ׀ H H H Unsaturated hydrocarbon (C = C double bond) H H H H H ׀ ׀ ׀ ׀ ׀ C = C – C – C – C - H ׀ ׀ ׀ ׀ H H H H ethene propene butene pentene
- 3. Class Functional group/name Examples alkene C = C Alkenyl ethene alkyne C ≡ C Alkynyl ethyne alcohol OH Hydroxyl ethanol ether C – O - C Ether methoxymethane ketone O ‖ C – C - C Carbonyl propanone aldehyde CHO Aldehyde ethanal Carboxylic acid COOH Carboxyl ethanoic acid ester O ‖ C – O -R Ester ethyl ethanoate amide O ‖ C – NH2 Amide propanamide amine NH2 Amine ethanamine nitrile C ≡ N Nitrile propanenitrile Class Functional gp Suffix Example Formula Alkane C - C - ane ethane CnH2n+2 Homologous Series carbon IUPAC name Structure formula Molecular formula Boiling point 1 Methane CH4 CH4 Gas 2 Ethane CH3CH3 C2H6 Gas 3 Propane CH3CH2CH3 C3H8 Gas 4 Butane CH3(CH2)2CH3 C4H10 Gas 5 Pentane CH3(CH2)3CH3 C5H12 Liquid 6 Hexane CH3(CH2)4CH3 C6H14 Liquid Physical properties • Increase RMM / molecular size •RMM increase ↑ - Van Der Waals forces stronger ↑ ↓ Melting /boiling point increases ↑ (Increasing polarisability ↑) London dispersion forces/temporary dipole ↑ 1 2 3 4 5 6 7 8 9 10 number carbons – RMM ↑ 150 100 50 0 -50 -100 -150 -200 b/p increase ↑ boiling point room temp gas liquid Homologous Series number Carbons / RMM ↑ 1 2 3 4 5 6 7 8 9 10 boiling point boiling point increase with increase carbon atoms alcohol alkane alkene alkyne London dispersion force (temporary dipole) H2 bonding carboxylic acid > alkane/alkene/alkyne alcohol carboxylic acid
- 4. Number carbon Molecular weight b/p Predicted poly fit 3rd order 1 2 3 4 5 72.15 36 6 86.18 69 7 100.21 98.4 8 114.23 125.6 9 128.2 151 10 142.28 174.1 11 156.31 196 12 170.33 216.2 13 184.37 234 14 198.39 253.6 15 212.42 270.6 16 226.41 286.9 17 240.4 302 18 254.5 317 19 268.5 330 20 282.5 343 21 296.6 363 22 310.6 368 23 324.6 379 24 338.6 391 25 352.7 403 IA secondary data based – Simple linear regression analysis for boiling point estimation Research Question Use 25 carbon chains for regression model Use regression eqn to estimate the b/p for 25 carbon chain Find the % error using expt values with predicted values. Using molecular weight 3rd order as estimator for b/p. Using carbon chain 3rd order as estimator for b/p. Using molecular weight 3rd order as estimator for b/p Number carbon Molecular weight b/p Predicted poly fit 3rd order 1 2 3 4 5 72.15 36 6 86.18 69 7 100.21 98.4 8 114.23 125.6 9 128.2 151 10 142.28 174.1 11 156.31 196 12 170.33 216.2 13 184.37 234 14 198.39 253.6 15 212.42 270.6 16 226.41 286.9 17 240.4 302 18 254.5 317 19 268.5 330 20 282.5 343 21 296.6 363 22 310.6 368 23 324.6 379 24 338.6 391 25 352.7 403 Using carbon chain 3rd order as estimator for b/p
- 5. Predicted b/p for carbon 10 – MW of 142.3 3rd order fit, y = 0.000006x3 – 0.0064x2 + 3.0855x – 153.76 b/p=0.000006(142.3)3 – 0.0064(142.3)2 + 3.0855(142.3) – 153.76 = 173 Research Question Which model, molecular weight model, 3rd order, better estimator for b/p. Which model, carbon chain model, 3rd order, better estimator for b/p. y = 6E-06x3 - 0.0064x2 + 3.0855x - 153.76 R² = 0.9998 0 50 100 150 200 250 300 350 400 450 0 50 100 150 200 250 300 350 400 b/p molecular weight molecular weight vs b/p y = 0.0171x3 - 1.2705x2 + 43.433x - 155.23 R² = 0.9992 0 50 100 150 200 250 300 350 400 450 0 5 10 15 20 25 30 b/p number of carbon chains number of carbon chains vs b/p Predicted b/p for carbon 10 3rd order fit, y = 0.0171x3 – 1.2705x2 + 43.433x – 155.23 b/p=0.0171(10)3 – 1.2705(10)2 + 43.433(10) – 155.23 =169 Predicted b/p for carbon 26 – MW of 366.7 3rd order fit, y = 0.000006x3 – 0.0064x2 + 3.0855x – 153.76 b/p=0.000006(366.7)3 – 0.0064(366.7)2 + 3.0855(366.7) – 153.76 = 413 Predicted b/p for carbon 30 – MW of 422.8 3rd order fit, y = 0.000006x3 – 0.0064x2 + 3.0855x – 153.76 b/p=0.000006(422.8)3 – 0.0064(422.8)2 + 3.0855(422.8) – 153.76 = 460 Predicted b/p for carbon 26 3rd order fit, y = 0.0171x3 – 1.2705x2 + 43.433x – 155.23 b/p=0.0171(26)3 – 1.2705(26)2 + 43.433(26) – 155.23 = 416 Predicted b/p for carbon 30 3rd order fit, y = 0.0171x3 – 1.2705x2 + 43.433x – 155.23 b/p=0.0171(30)3 – 1.2705(30)2 + 43.433(30) – 155.23 = 466 Using molecular weight 3rd order as estimator for b/p Using carbon chain 3rd order as estimator for b/p
- 6. Using molecular weight 3rd order as estimator for b/p Using carbon chain 3rd order as estimator for b/p Number carbon Molecular weight b/p predicted poly fit 3rd order (% error) 1 2 3 4 5 72.15 36 6 86.18 69 7 100.21 98.4 8 114.23 125.6 9 128.2 151 10 142.28 174.1 173 (0.5%) 11 156.31 196 12 170.33 216.2 13 184.37 234 14 198.39 253.6 15 212.42 270.6 16 226.41 286.9 17 240.4 302 18 254.5 317 19 268.5 330 20 282.5 343 21 296.6 363 22 310.6 368 23 324.6 379 24 338.6 391 25 352.7 403 26 366.7 412 413 (0.2%) 27 378.8 422 28 392.7 431 29 406.8 440 30 422.8 449.7 460 (2%) Number carbon Boiling point predicted poly fit 3rd order (% error) 1 2 3 4 5 36 6 69 7 98.4 8 125.6 9 151 10 174.1 169 (2.9%) 11 196 12 216.2 13 234 14 253.6 15 270.6 16 286.9 17 302 18 317 19 330 20 343 21 363 22 368 23 379 24 391 25 403 26 412 416 (1%) 27 422 28 431 29 440 30 449.7 466 (3.6%) % error = (𝑬𝒙𝒑𝒕 𝒗𝒂𝒍𝒖𝒆 −𝑷𝒓𝒆𝒅𝒊𝒄𝒕𝒆𝒅 𝒗𝒂𝒍𝒖𝒆) 𝑬𝒙𝒑𝒕 𝒗𝒂𝒍𝒖𝒆 x 100% % error = (𝟏𝟕𝟒.𝟏 −𝟏𝟕𝟑) 𝟏𝟕𝟒.𝟏 x 100% = 0.5% % error = (𝑬𝒙𝒑𝒕 𝒗𝒂𝒍𝒖𝒆 −𝑷𝒓𝒆𝒅𝒊𝒄𝒕𝒆𝒅 𝒗𝒂𝒍𝒖𝒆) 𝑬𝒙𝒑𝒕 𝒗𝒂𝒍𝒖𝒆 x 100% % error = (𝟏𝟕𝟒.𝟏 −𝟏𝟔𝟗) 𝟏𝟕𝟒.𝟏 x 100% = 2.9%
- 7. Research Question Use regression eqn to estimate the b/p for 10, 26 and 30 based on molecular weight. Which model, molecular weight model, better estimator for b/p. Which model, carbon chain model, better estimator for b/p. Result showed molecular weight model is a better fit % error, smaller compared to carbon chain model % error increases as molecular weight increases 3rd order fit – % error changes from 0.5% to 0.2% to 2% as carbon chain changes from 10 to 26 to 30 y = 6E-06x3 - 0.0064x2 + 3.0855x - 153.76 R² = 0.9998 0 50 100 150 200 250 300 350 400 450 0 50 100 150 200 250 300 350 400 b/p molecular weight molecular weight vs b/p Number carbon Molecular weight b/p predicted poly fit 3rd order (% error) 10 142.28 174.1 173 (0.5%) 26 366.7 412 413 (0.2%) 30 422.8 449.7 460 (2%) y = 0.0171x3 - 1.2705x2 + 43.433x - 155.23 R² = 0.9992 0 50 100 150 200 250 300 350 400 450 0 5 10 15 20 25 30 b/p number of carbon chains number of carbon chains vs b/p Number of carbon b/p predicted poly fit 3rd order (% error) 10 174.1 169 (2.9%) 26 412 416 (1%) 30 449.7 466 (3.6%) Result showed carbon chain model is a weaker fit % error, bigger compared to molecular weight model. % error increases as carbon chains increases 3rd order fit – % error changes from 2.9% to 1% to 3.6% as carbon chain changes from 10 to 26 to 30
- 8. Predicted b/p for carbon 35 – MW of 493 3rd order fit, y = 0.000006x3 – 0.0064x2 + 3.0855x – 153.76 b/p=0.000006(493)3 – 0.0064(493)2 + 3.0855(493) – 153.76 = 529 Polynomial 3rd order molecular weight model Number carbon Molecular weight b/p predicted poly fit 3rd order 5 72.15 36 6 86.18 69 7 100.21 98.4 8 114.23 125.6 9 128.2 151 10 142.28 174.1 173 26 366.7 412 413 30 422.8 449.7 460 35 493 490 529 y = 6E-06x3 - 0.0064x2 + 3.0855x - 153.76 R² = 0.9998 0 200 400 600 0 50 100 150 200 250 300 350 400 b/p molecular weight molecular weight vs b/p Number carbon Molecular weight b/p predicted poly fit 3rd order (% error) 10 142.28 174.1 173 (0.5%) 26 366.7 412 413 (0.2%) 30 422.8 449.7 460 (2%) 35 493 490 529 (8%) Polynomial 3rd order carbon chain model Number of carbon Boiling point Predicted poly fit 3rd order 5 36 6 69 7 98.4 8 125.6 9 151 10 174.1 169 26 412 416 30 449.7 466 35 490 541 Predicted b/p for carbon 35 3rd order fit, y = 0.0171x3 – 1.2705x2 + 43.433x – 155.23 b/p = 0.0171(35)3 – 1.2705(35)2 + 43.433(35) – 155.23 = 541 y = 0.0171x3 - 1.2705x2 + 43.433x - 155.23 R² = 0.9992 0 200 400 600 0 5 10 15 20 25 30 b/p number of carbon chains number of carbon chains vs b/p Number carbon b/p predicted poly fit 3rd order (% error) 10 174.1 169 (2.9%) 26 412 416 (1%) 30 449.7 466 (3.6%) 35 493 541 (10%)
- 9. Predicted b/p for carbon 35 – MW of 493 3rd order fit, y = 0.000006x3 – 0.0064x2 + 3.0855x – 153.76 b/p=0.000006(493)3 – 0.0064(493)2 + 3.0855(493) – 153.76 = 529 Number carbon Molecular weight b/p predicted poly fit 3rd order 5 72.15 36 6 86.18 69 7 100.21 98.4 8 114.23 125.6 9 128.2 151 10 142.28 174.1 173 26 366.7 412 413 30 422.8 449.7 460 35 493 490 529 Number carbon Molecular weight b/p predicted poly fit 3rd order (% error) 10 142.28 174.1 173 (0.5%) 26 366.7 412 413 (0.2%) 30 422.8 449.7 460 (2%) 35 493 490 529 (8%) % error increases as molecular weight increases 3rd order fit – % error changes from 0.5% to 0.2% to 2% to 8% as carbon chain changes from 10 to 26 to 30 to 35. Polynomial 3rd order molecular weight model Number carbon b/p predicted poly fit 3rd order 5 36 6 69 7 98.4 8 125.6 9 151 10 174.1 169 26 412 416 30 449.7 466 35 490 541 Polynomial 3rd order carbon chain model Predicted b/p for carbon 35 3rd order fit, y = 0.0171x3 – 1.2705x2 + 43.433x – 155.23 b/p = 0.0171(35)3 – 1.2705(35)2 + 43.433(35) – 155.23 = 541 Number carbon b/p predicted poly fit 3rd order (% error) 10 174.1 169 (2.9%) 26 412 416 (1%) 30 449.7 466 (3.6%) 35 493 541 (10%) % error increases as carbon chains increases 3rd order fit – % error changes from 2.9% to 1% to 3.6% to 10% as carbon chain changes from 10 to 26 to 30 to 35.
