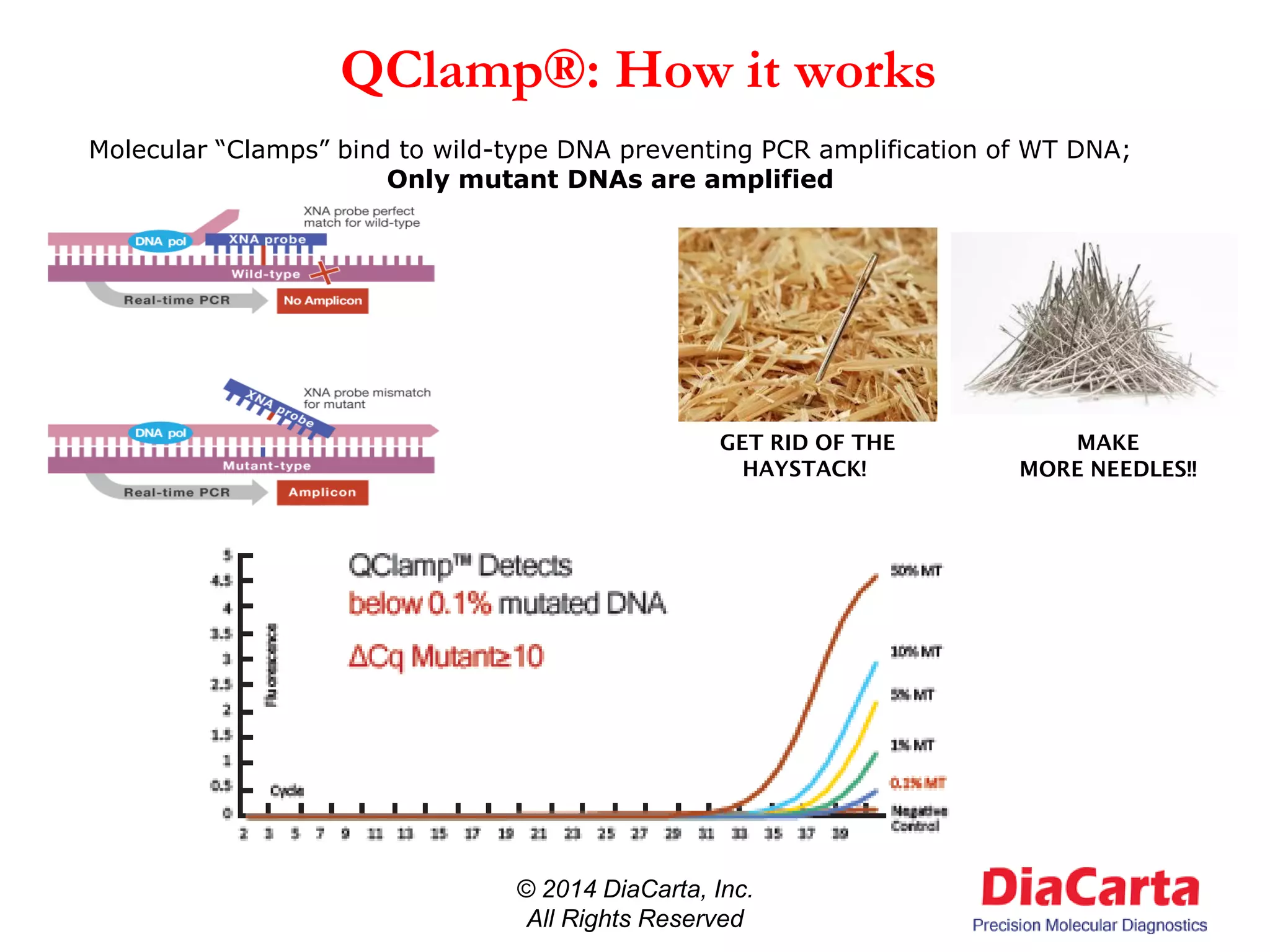
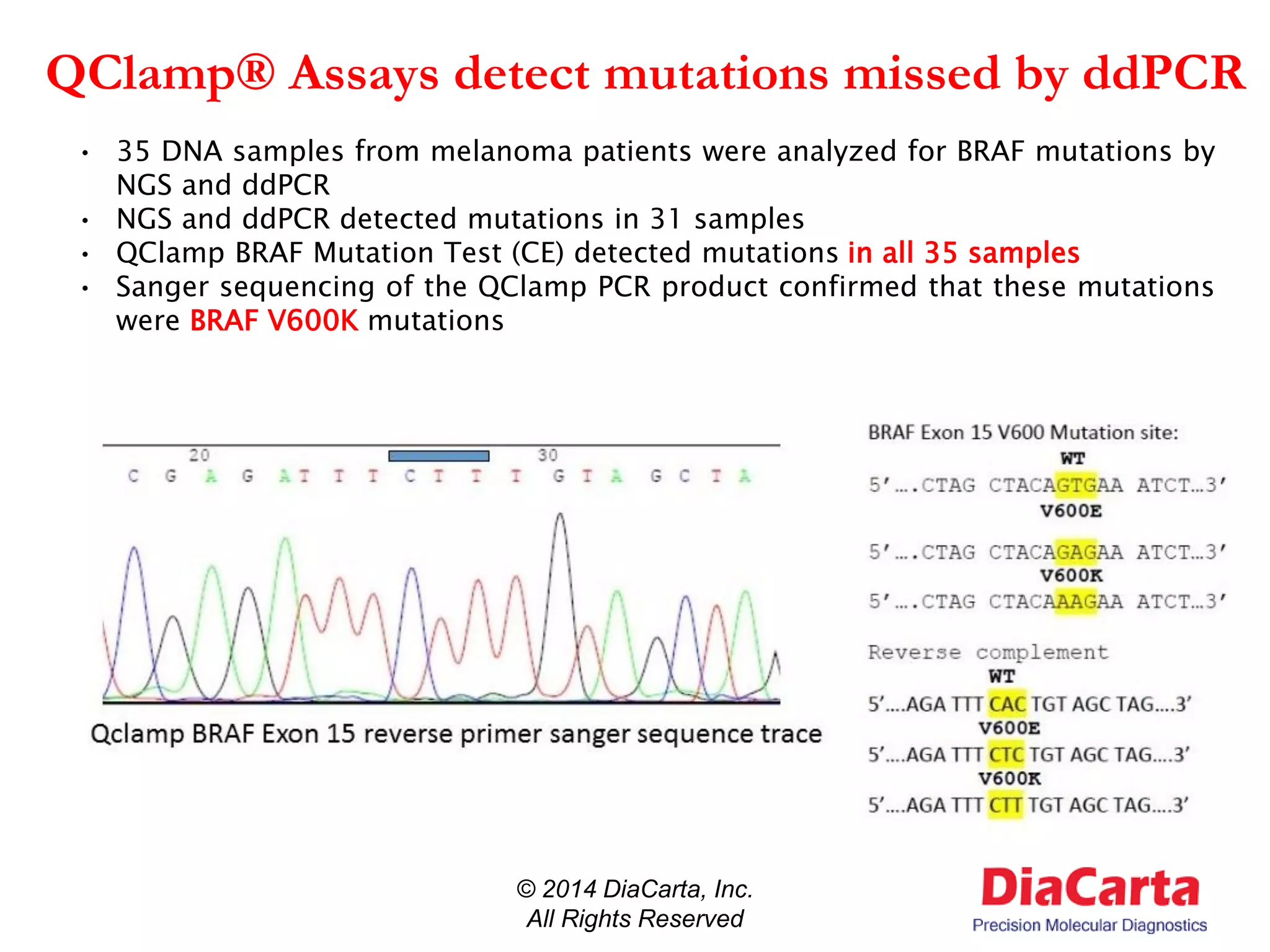
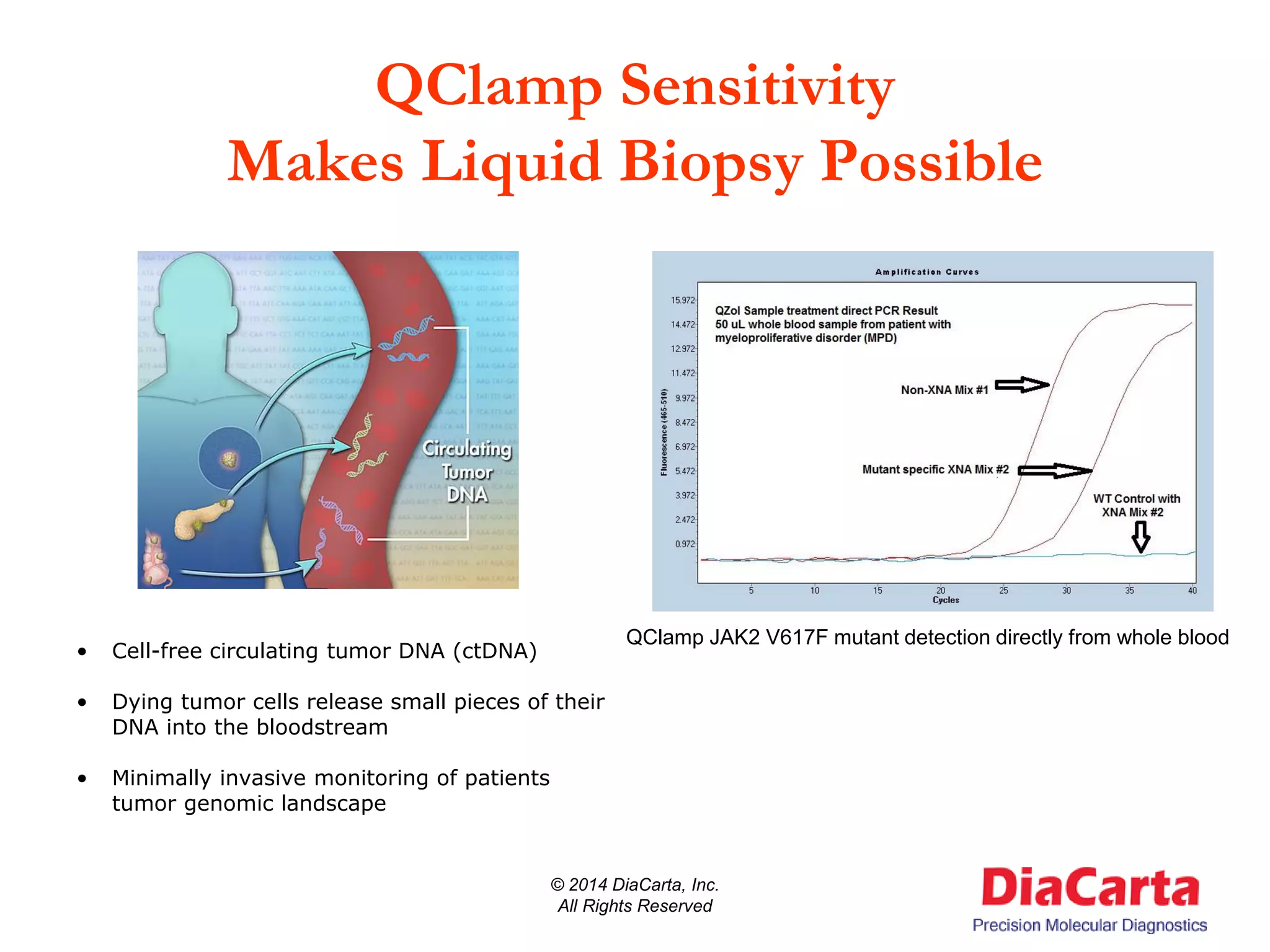
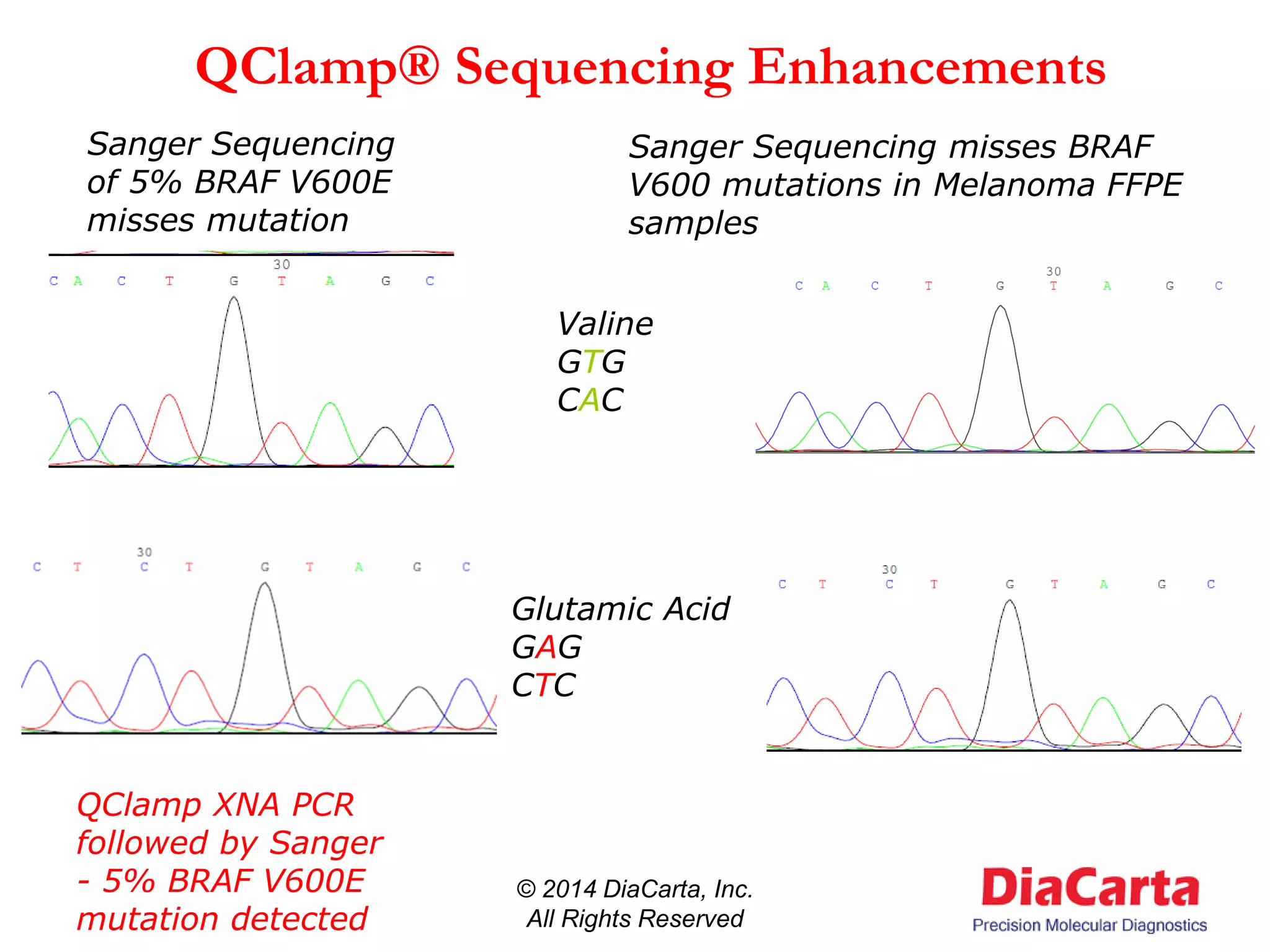
1) The document describes QClamp XNA-PCR, a technology from DiaCarta that uses molecular "clamps" to highly sensitively detect low frequency and cell-free circulating mutant DNA.
2) It needs assays that can detect biomarkers present at low frequency in samples containing a high amount of normal DNA. Existing technologies are inadequate for this need.
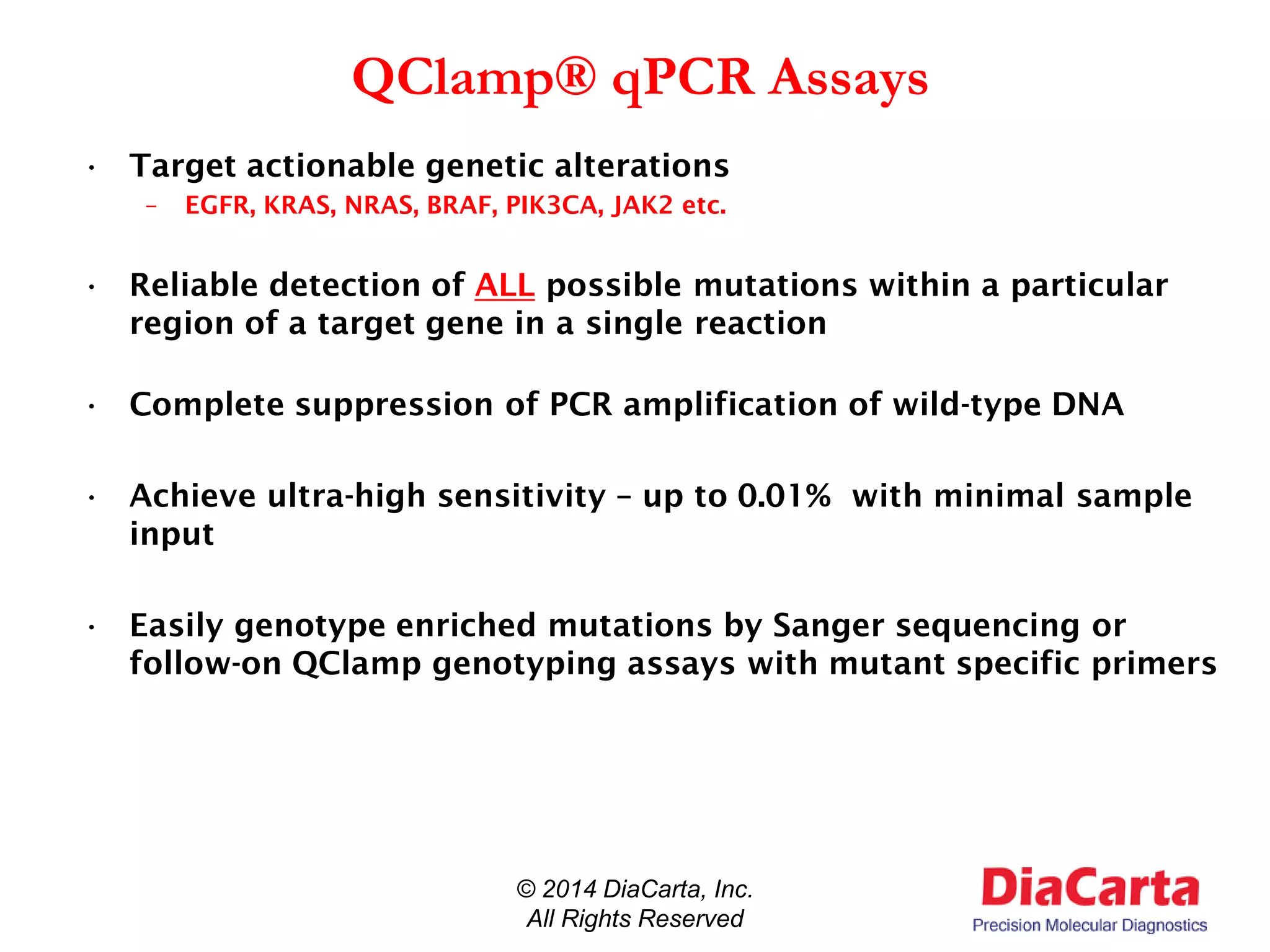
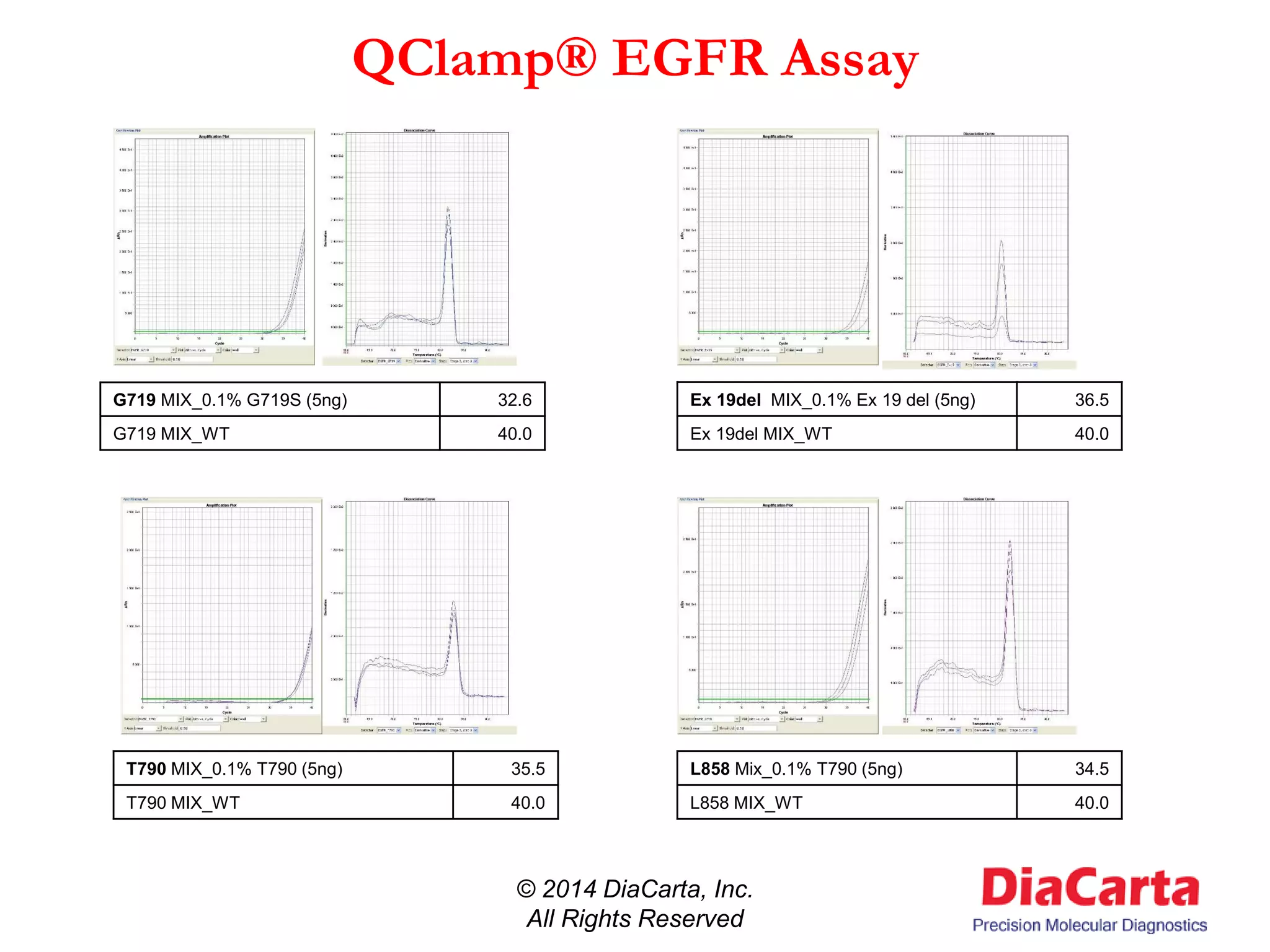
3) QClamp assays can reliably detect all possible mutations in a gene region using a single reaction. They can achieve ultra-high sensitivity down to 0.01% from minimal sample input.