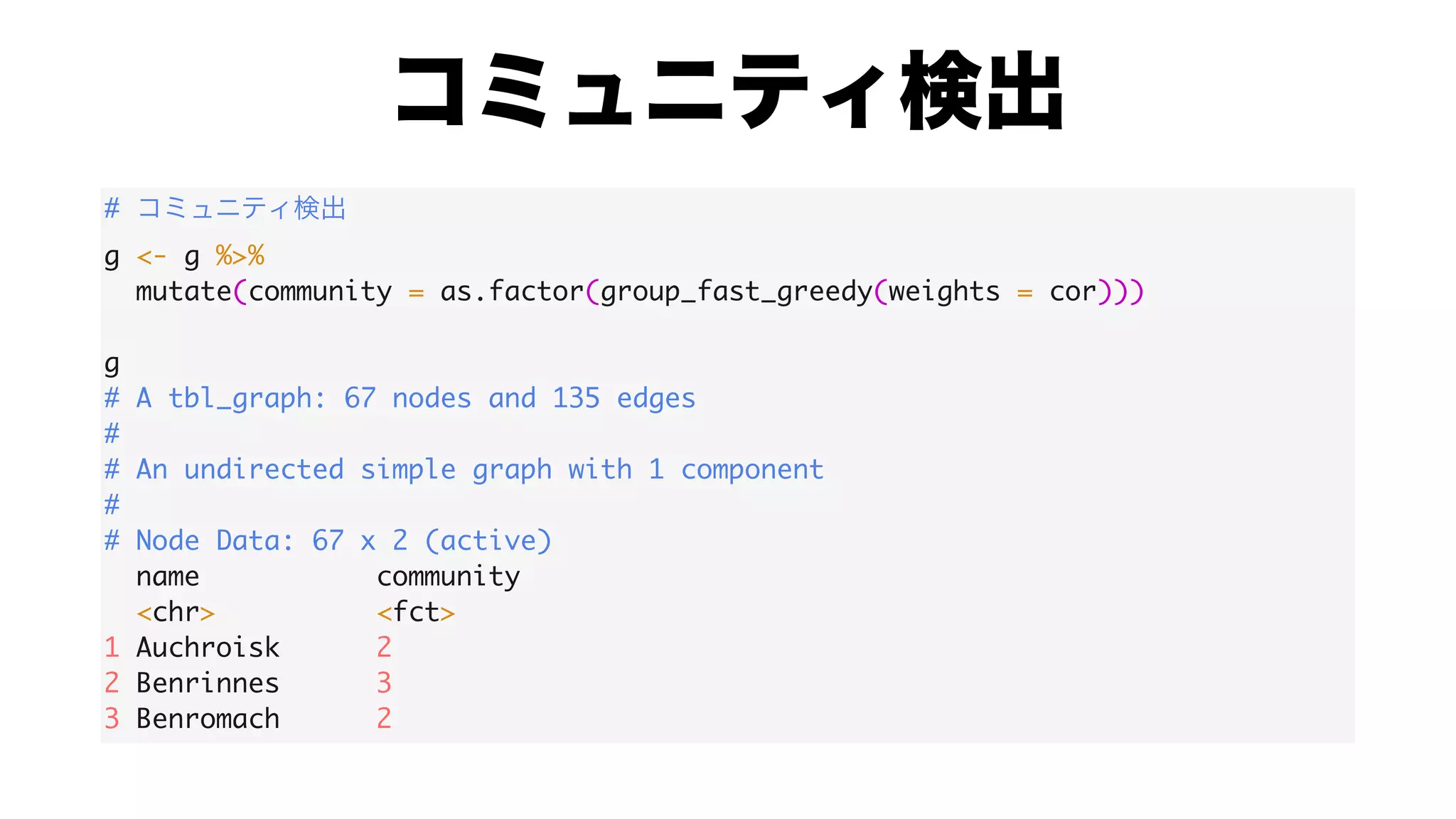
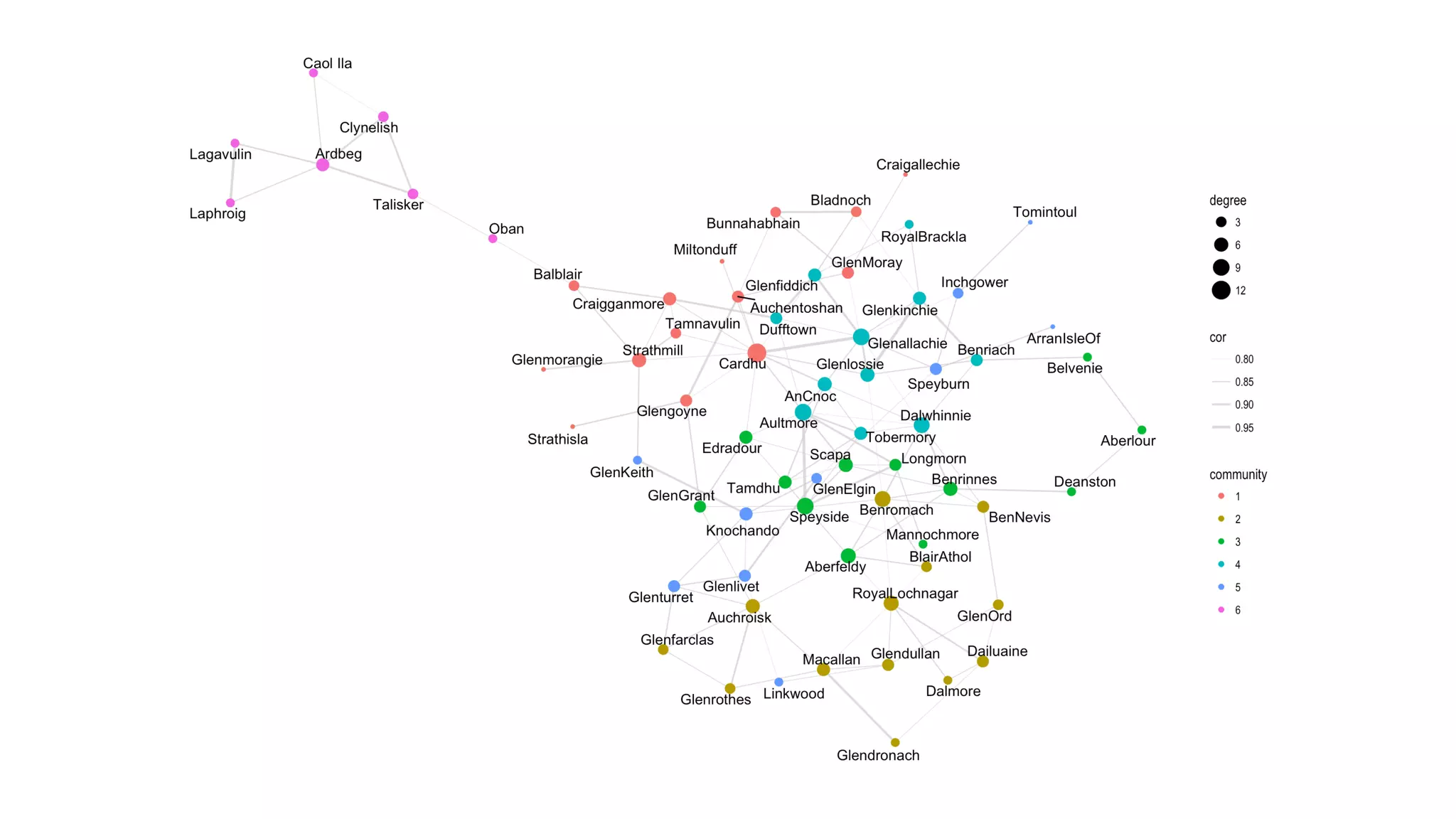
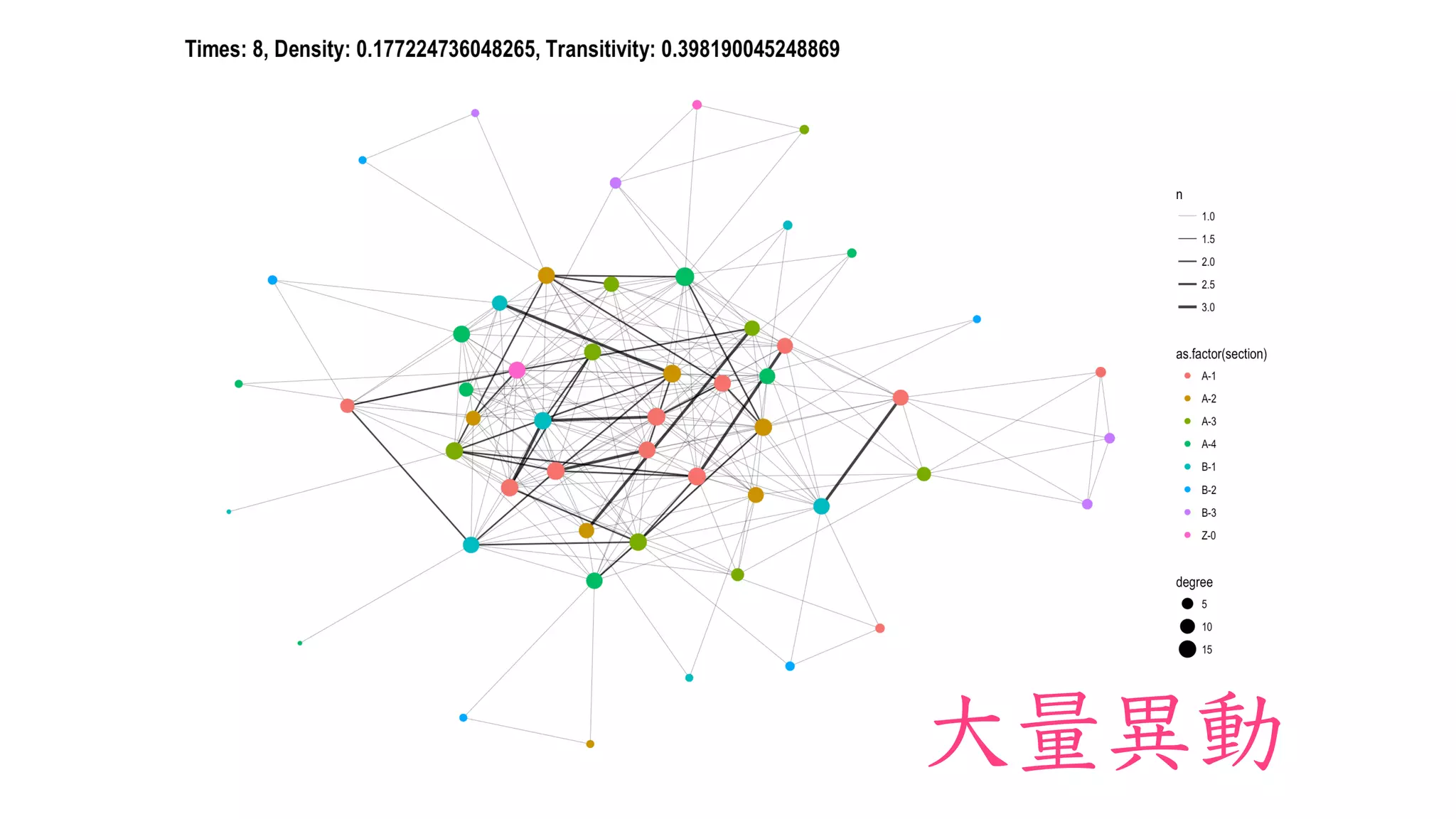
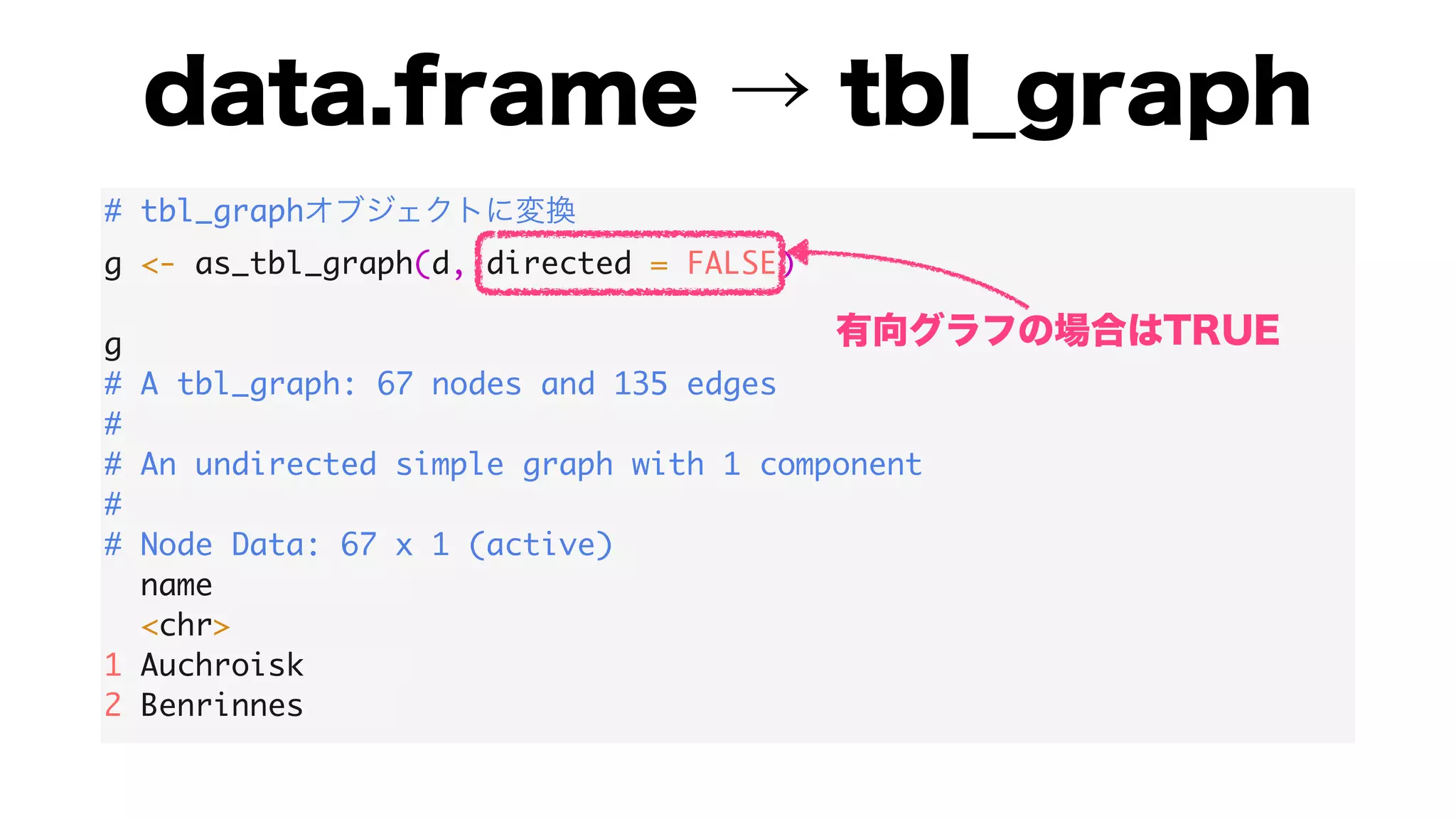
The document discusses analyzing correlation networks between Scottish whisky distilleries. A correlation matrix is created from sensory characteristics of whiskies. This is converted to a graph object where nodes are distilleries and edges represent correlations above 0.8. The graph is analyzed to find clustering of distilleries based on sensory profiles and key central nodes. Visualizations of the network are also created.

![> me
$name
[1] "Takashi Kitano"
$twitter
[1] "@kashitan"
$work_in
[1] " "](https://image.slidesharecdn.com/20180421tokyor69full-190628110634/75/tidygraph-ggraph-ver-2-2048.jpg)

























![#
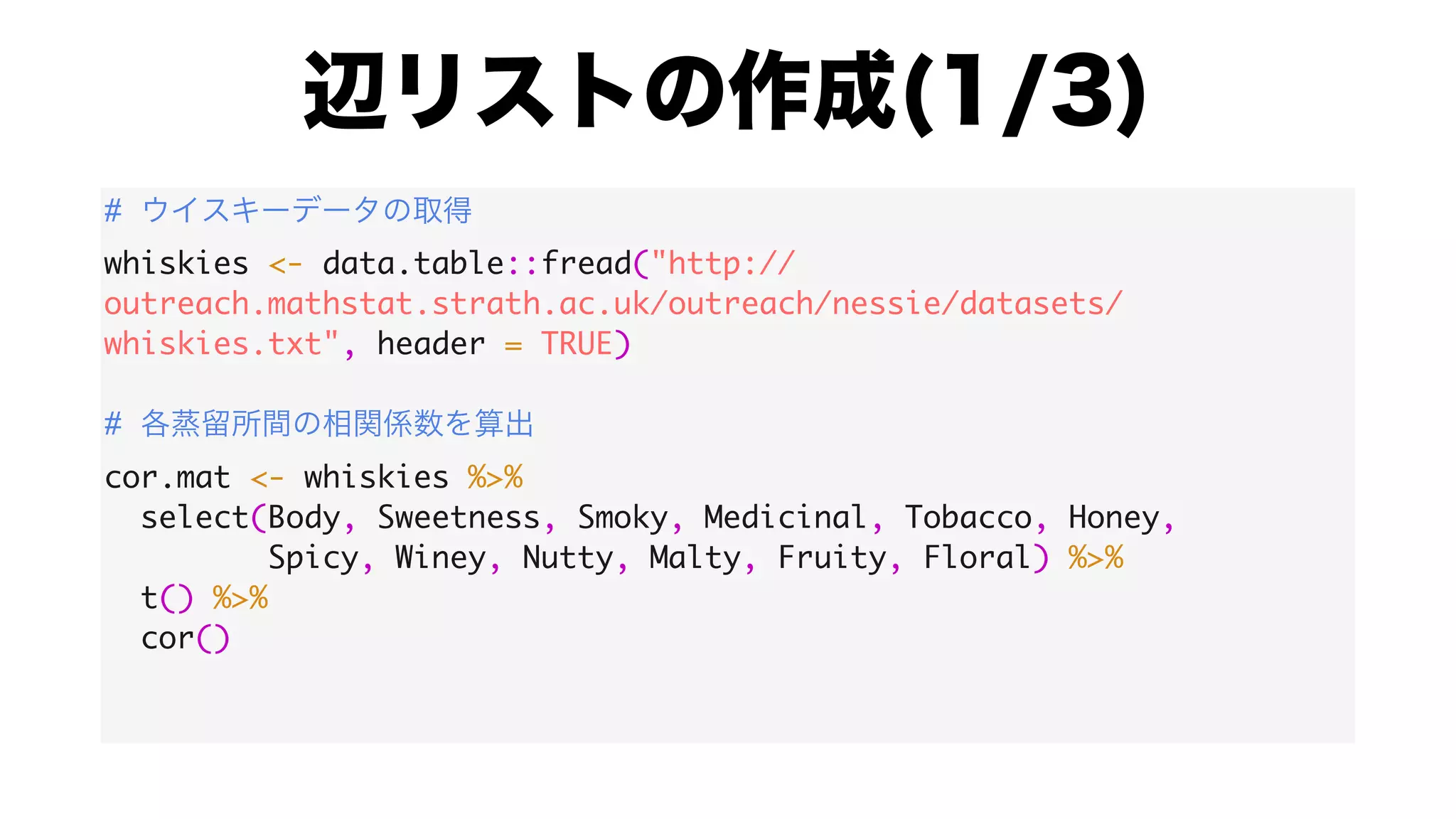
colnames(cor.mat) <- whiskies$Distillery
rownames(cor.mat) <- whiskies$Distillery
#
cor.mat[upper.tri(cor.mat, diag = TRUE)] <- NA
cor.mat[1:5, 1:5]
Aberfeldy Aberlour AnCnoc Ardbeg Ardmore
Aberfeldy NA NA NA NA NA
Aberlour 0.7086322 NA NA NA NA
AnCnoc 0.6973541 0.5030737 NA NA NA
Ardbeg -0.1473114 -0.2285909 -0.1404355 NA NA
Ardmore 0.7319024 0.5118338 0.5570195 0.2316174 NA](https://image.slidesharecdn.com/20180421tokyor69full-190628110634/75/tidygraph-ggraph-ver-31-2048.jpg)





![#
g %>% igraph::graph.density()
[1] 0.06105834
#
g %>% igraph::transitivity()
[1] 0.2797927
# ( 1)
g %>% igraph::reciprocity()
[1] 1](https://image.slidesharecdn.com/20180421tokyor69full-190628110634/75/tidygraph-ggraph-ver-38-2048.jpg)