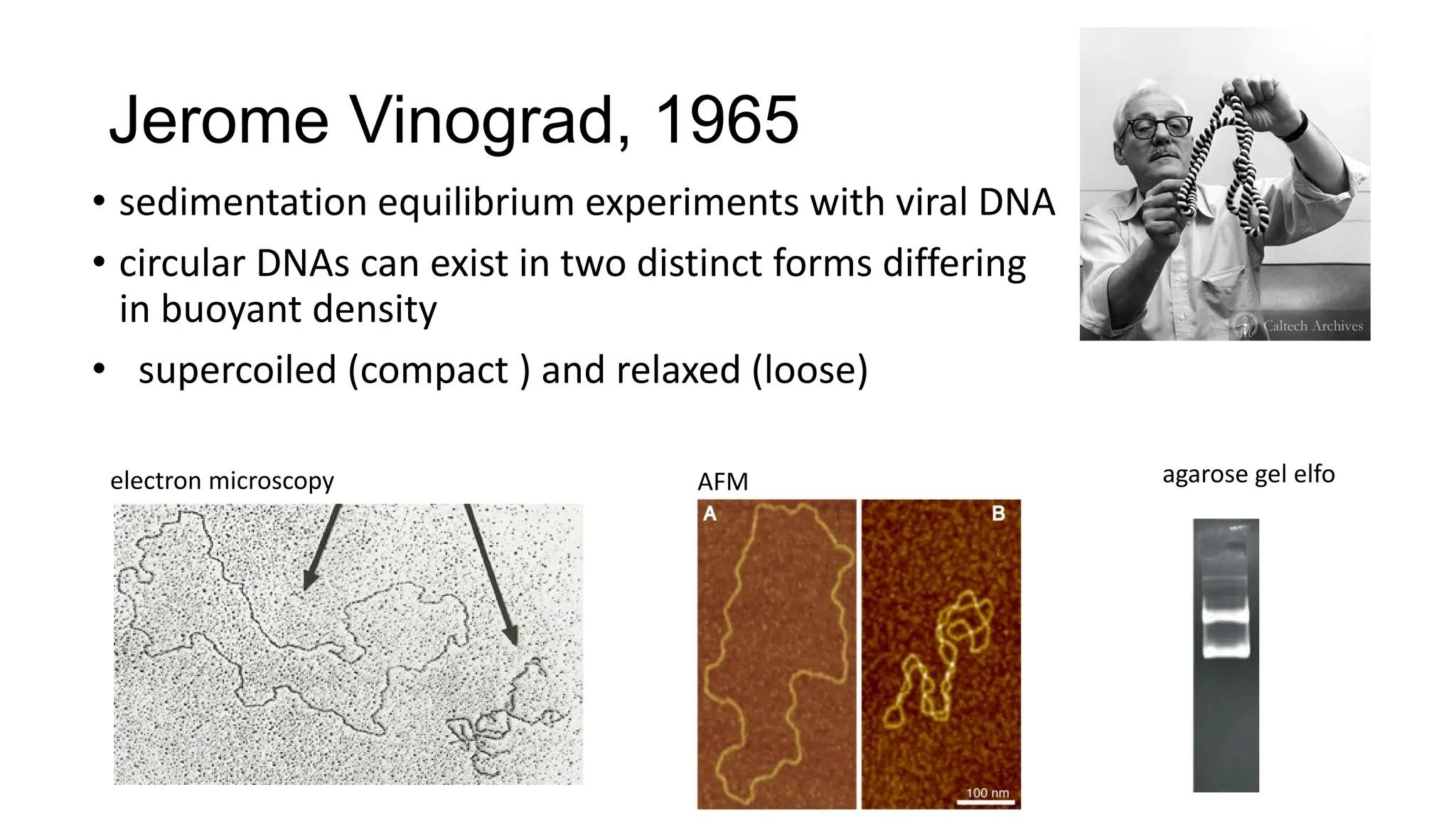
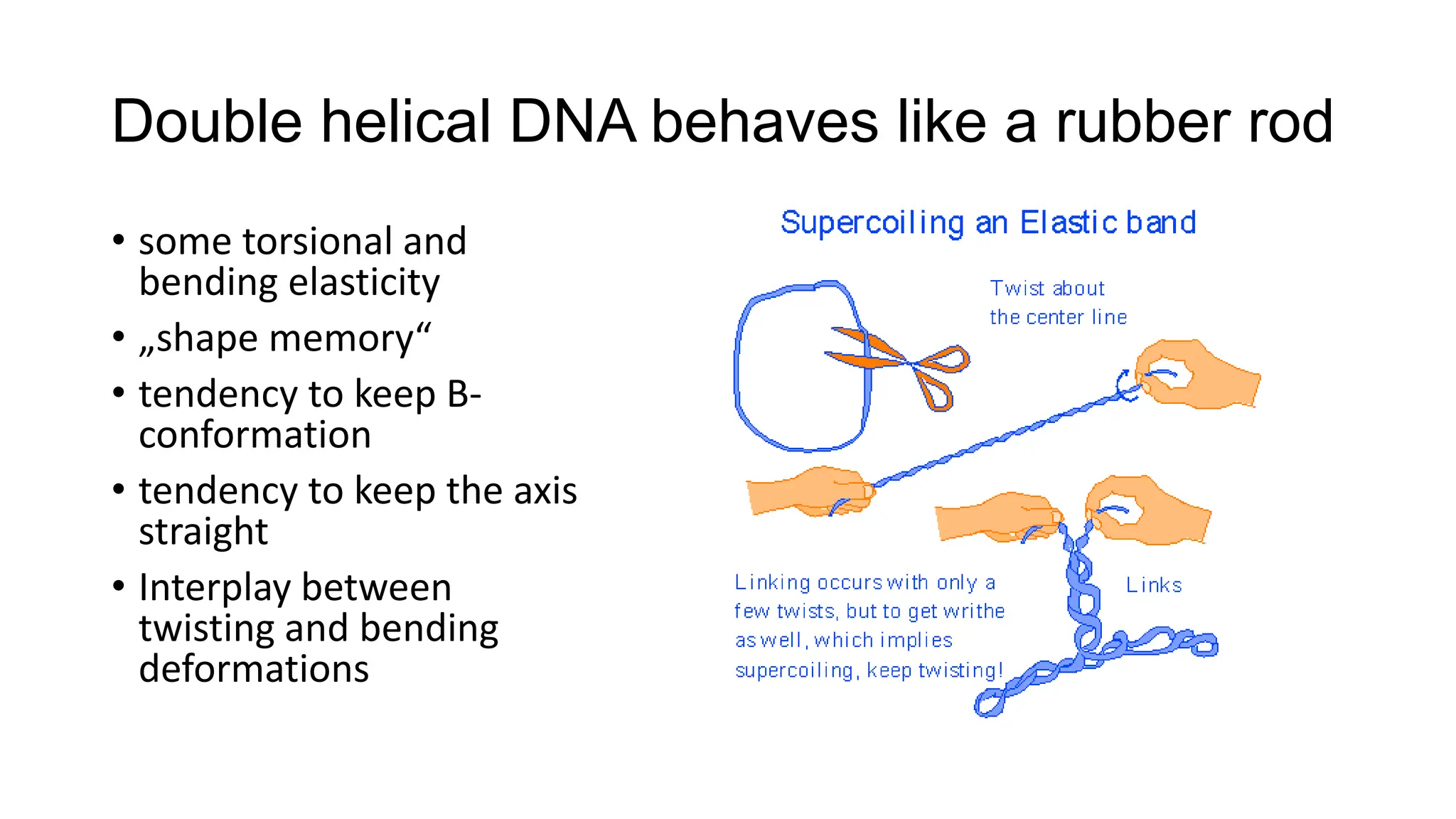
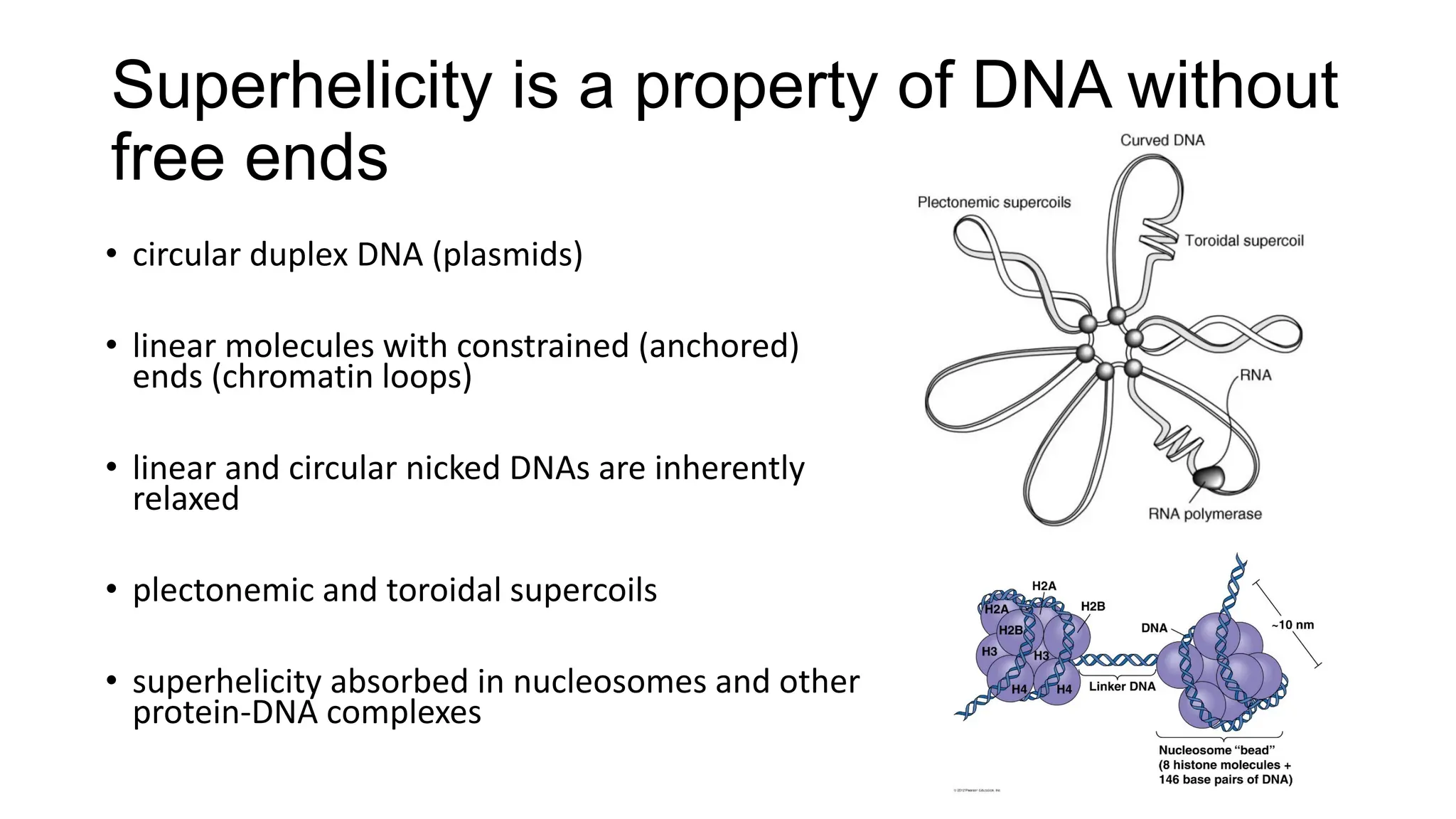
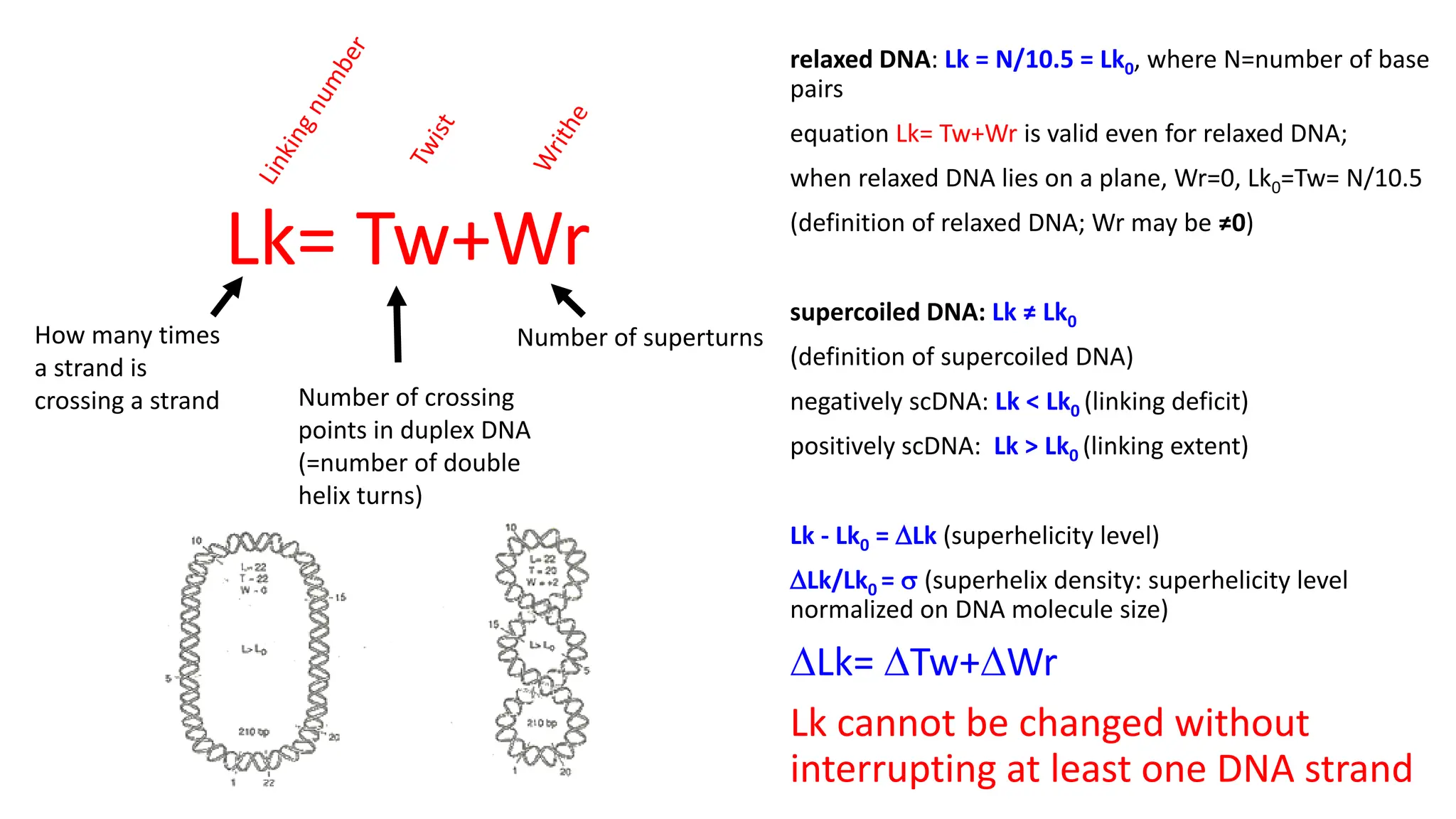
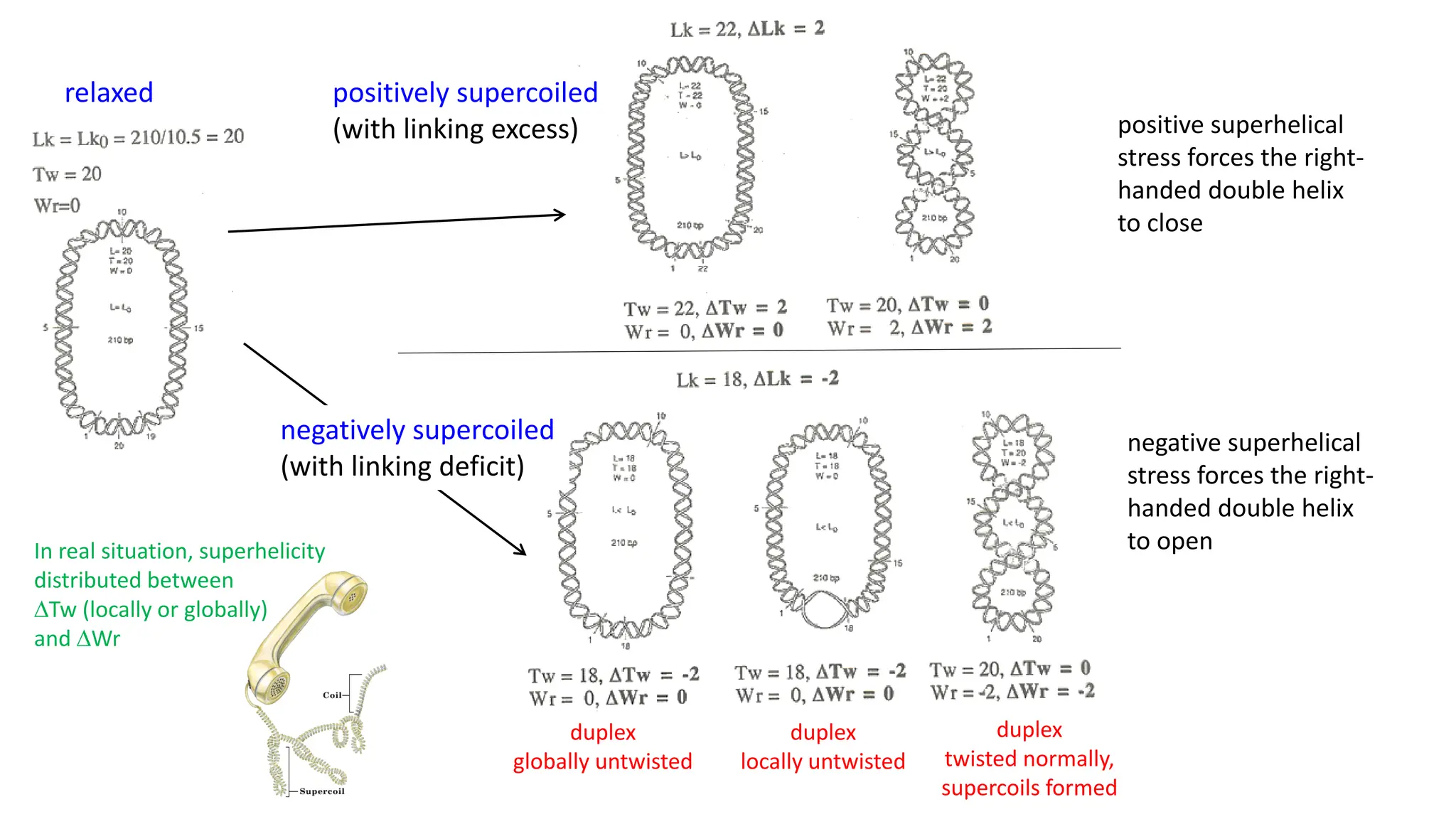
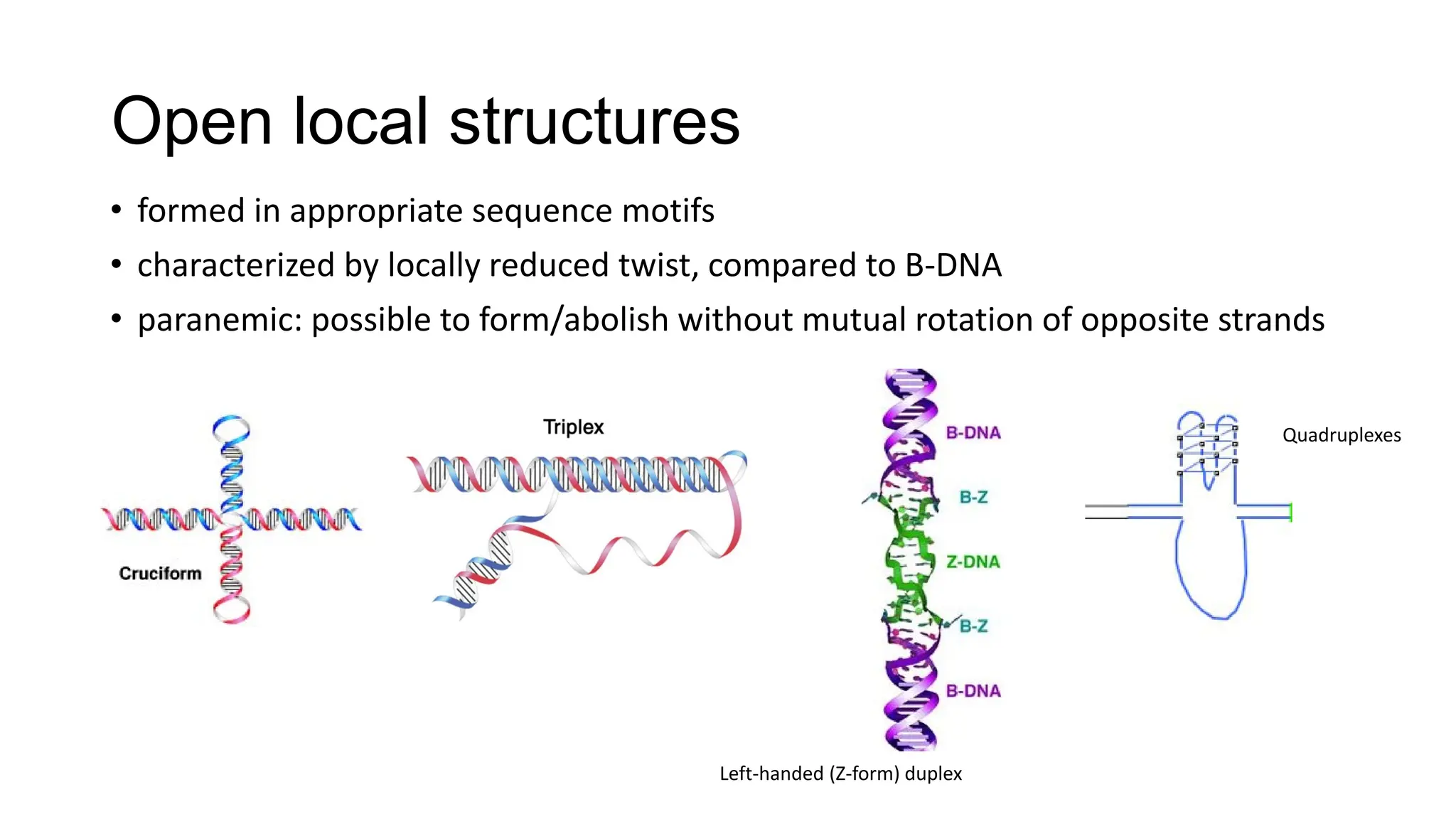
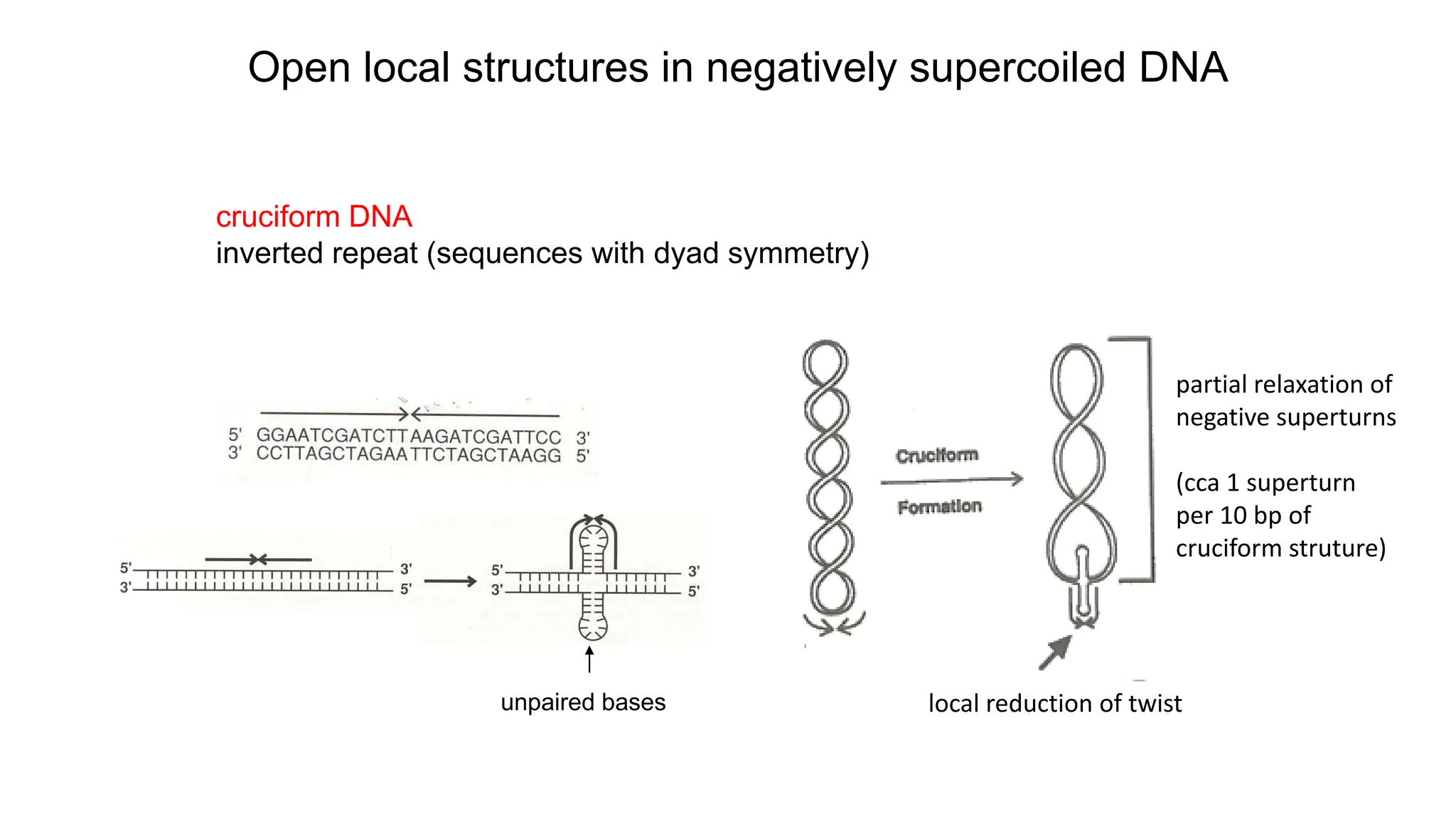
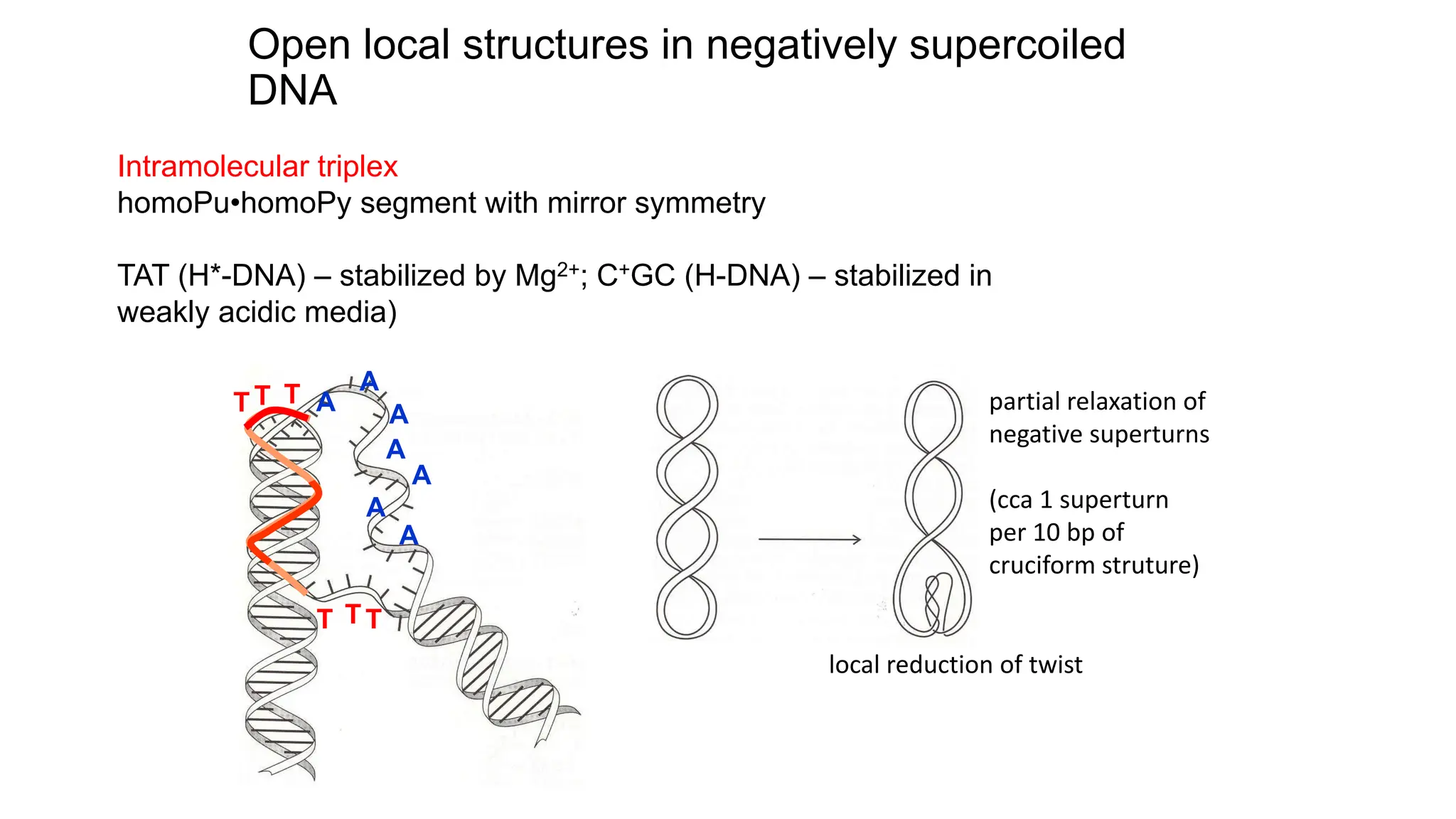
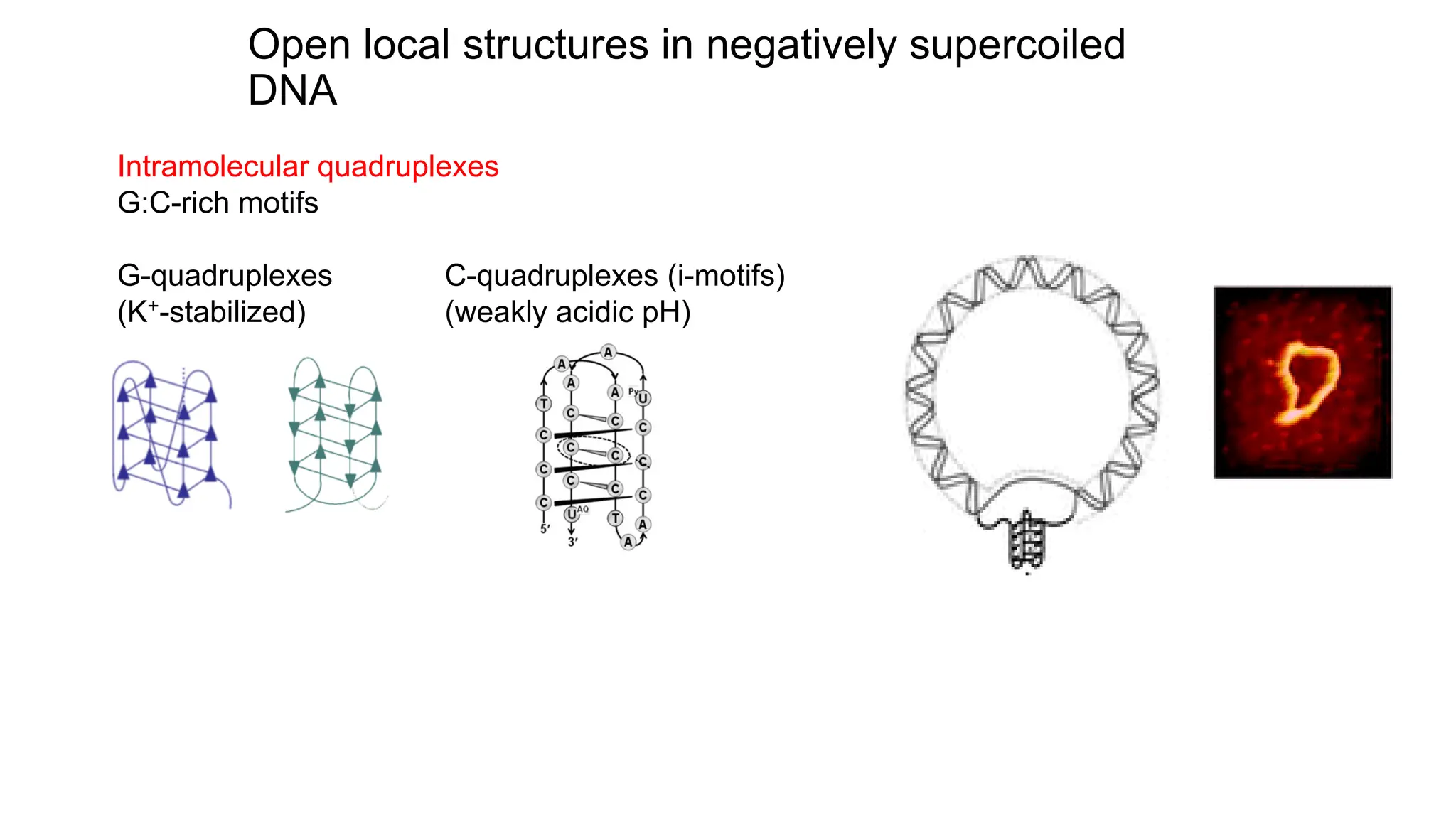
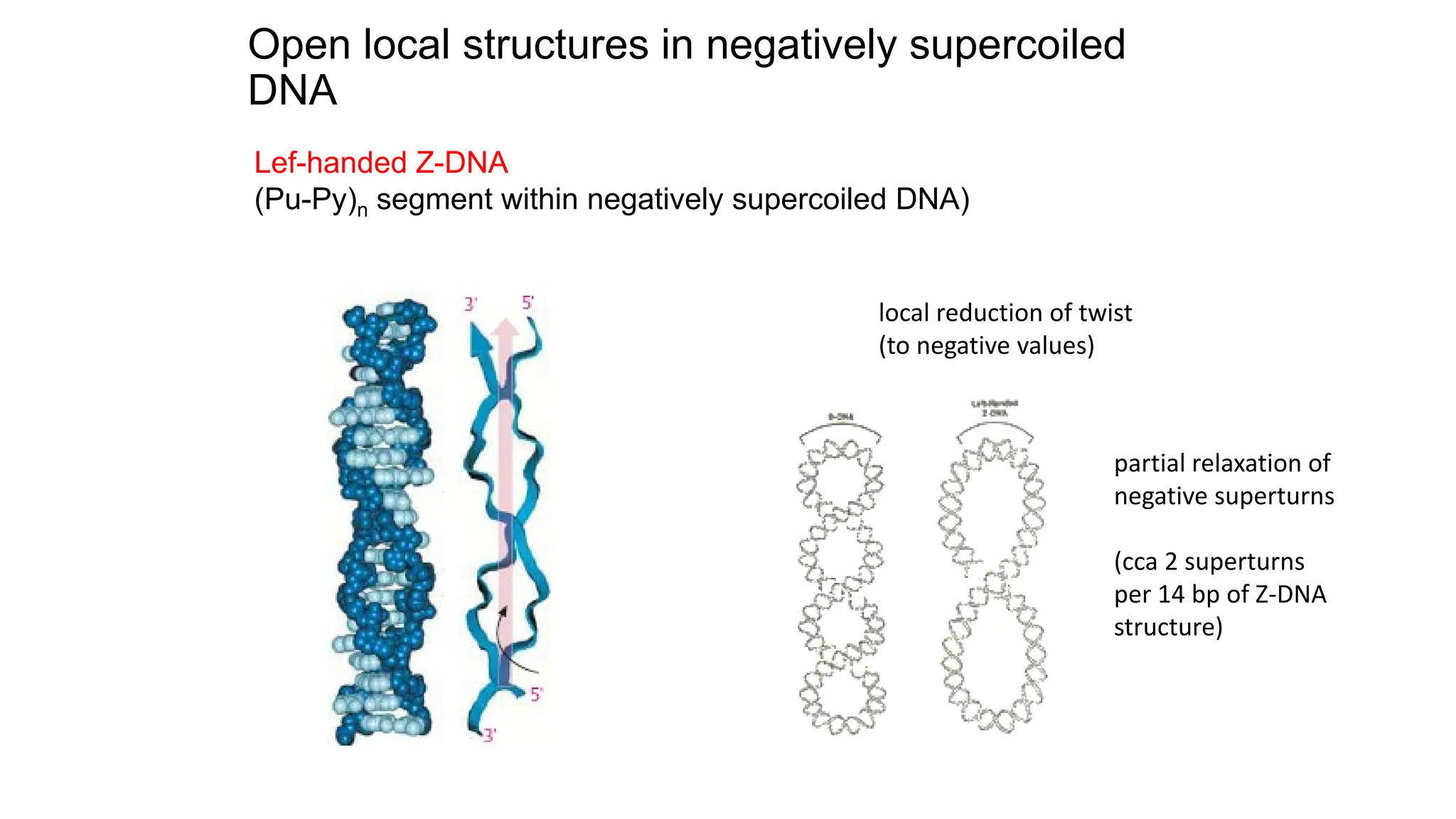
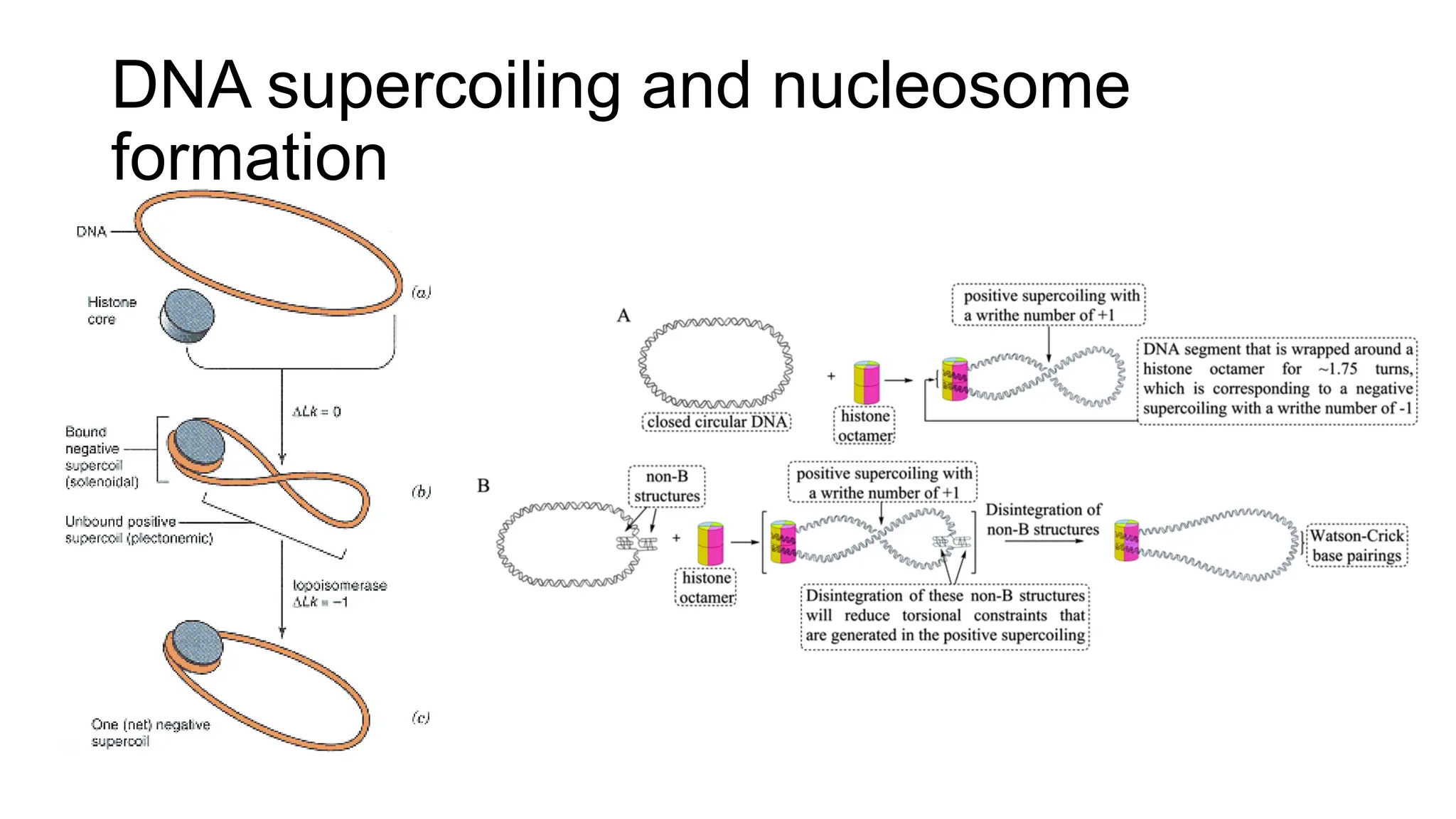
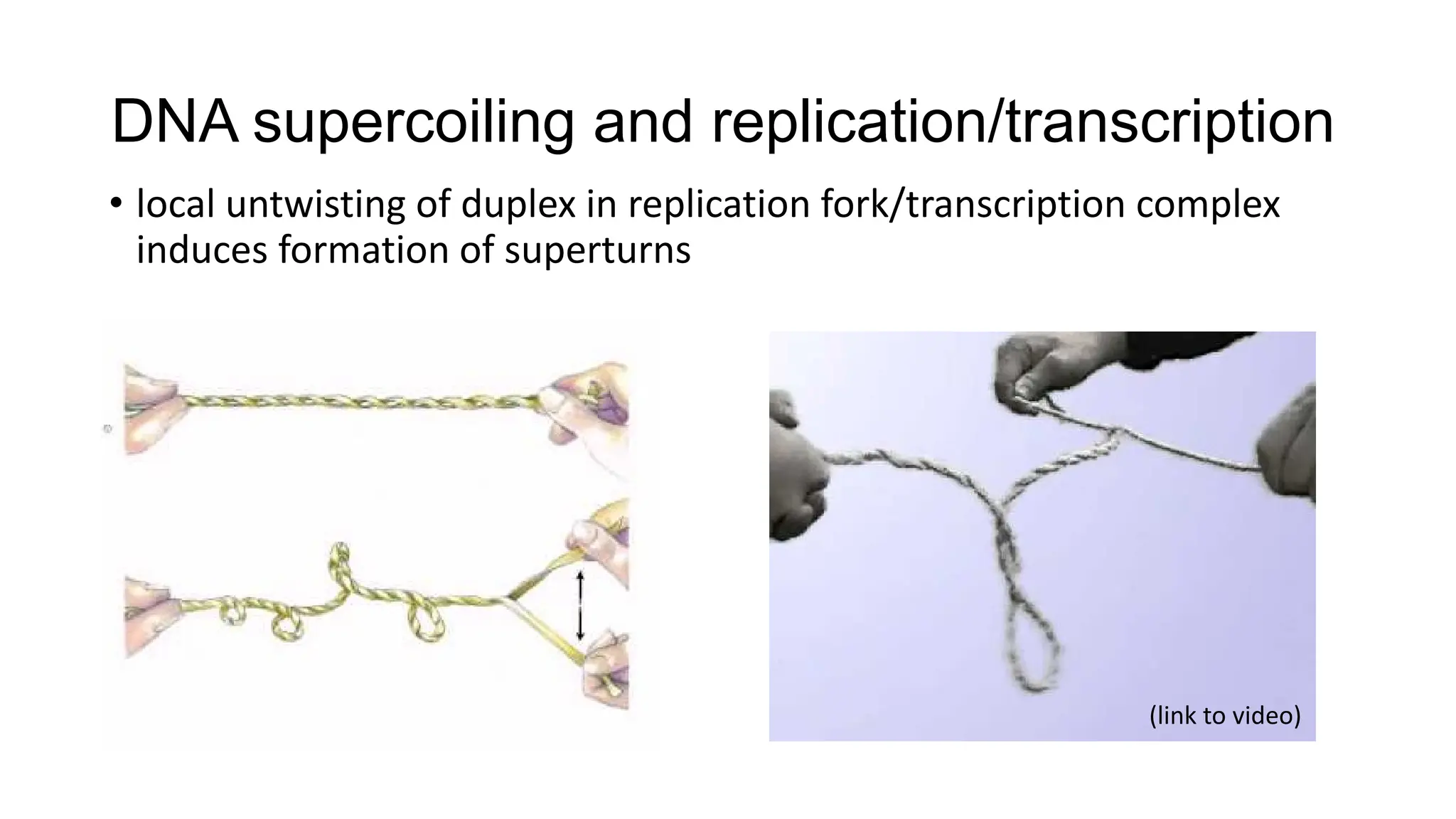
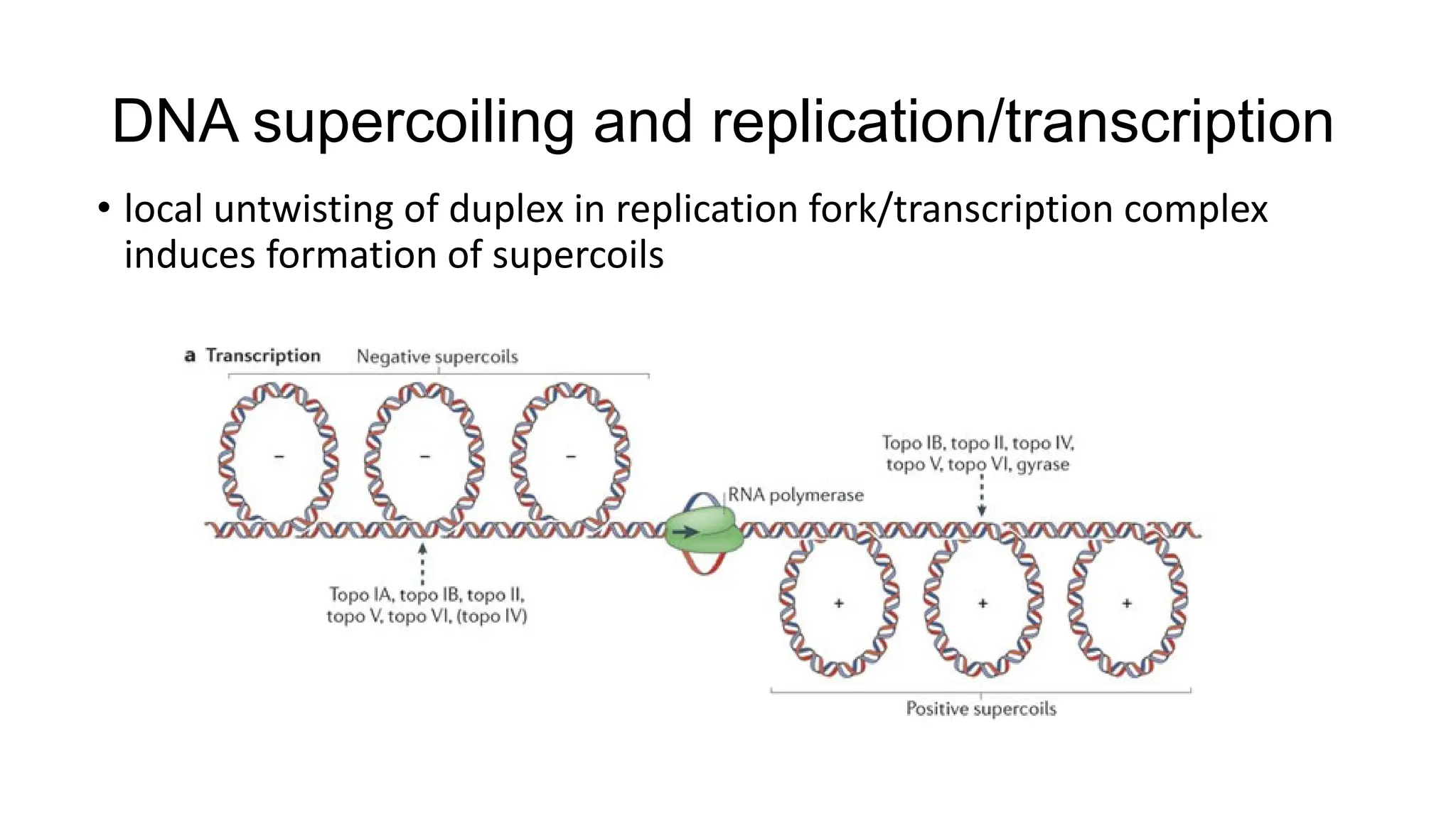
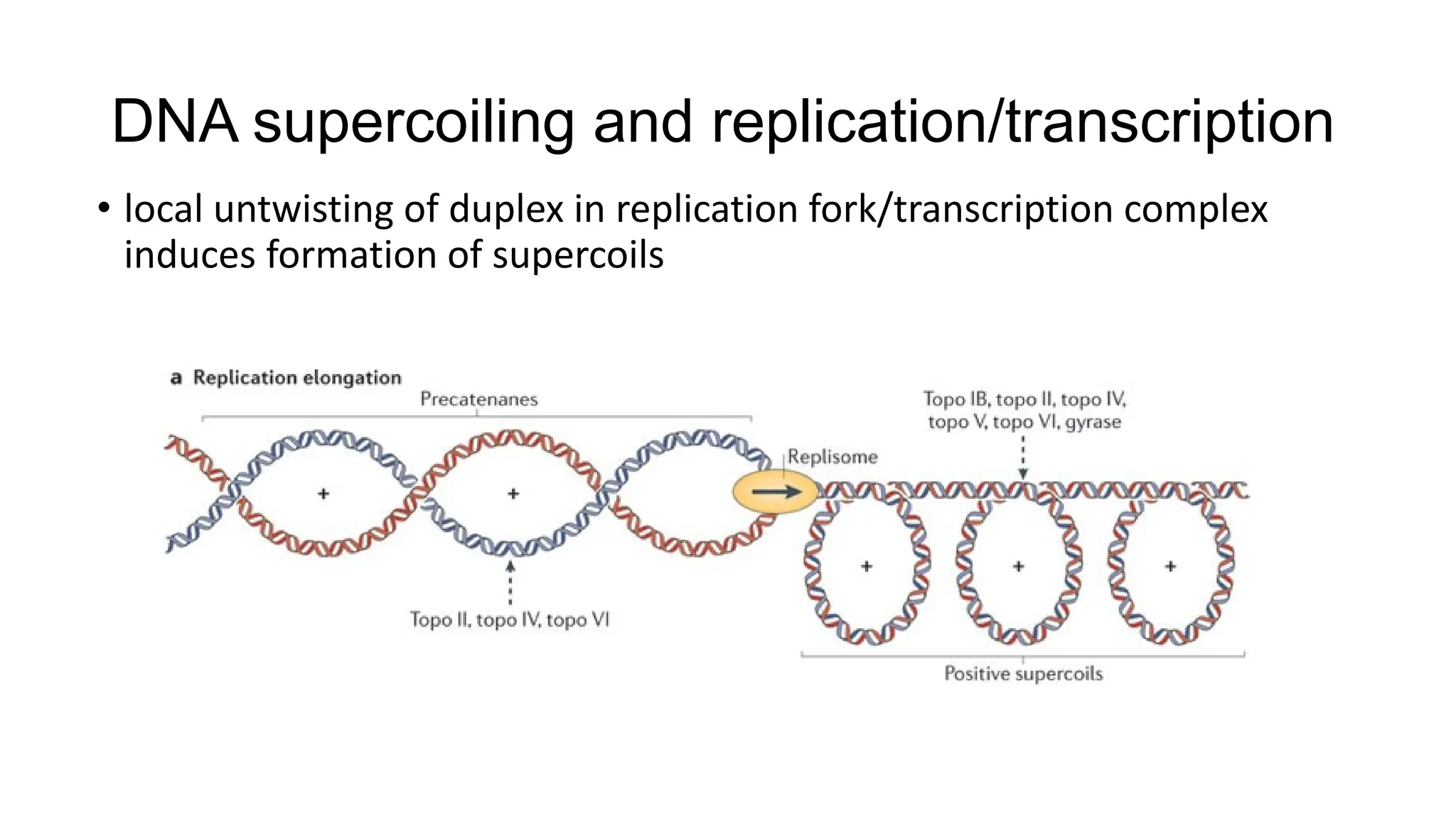
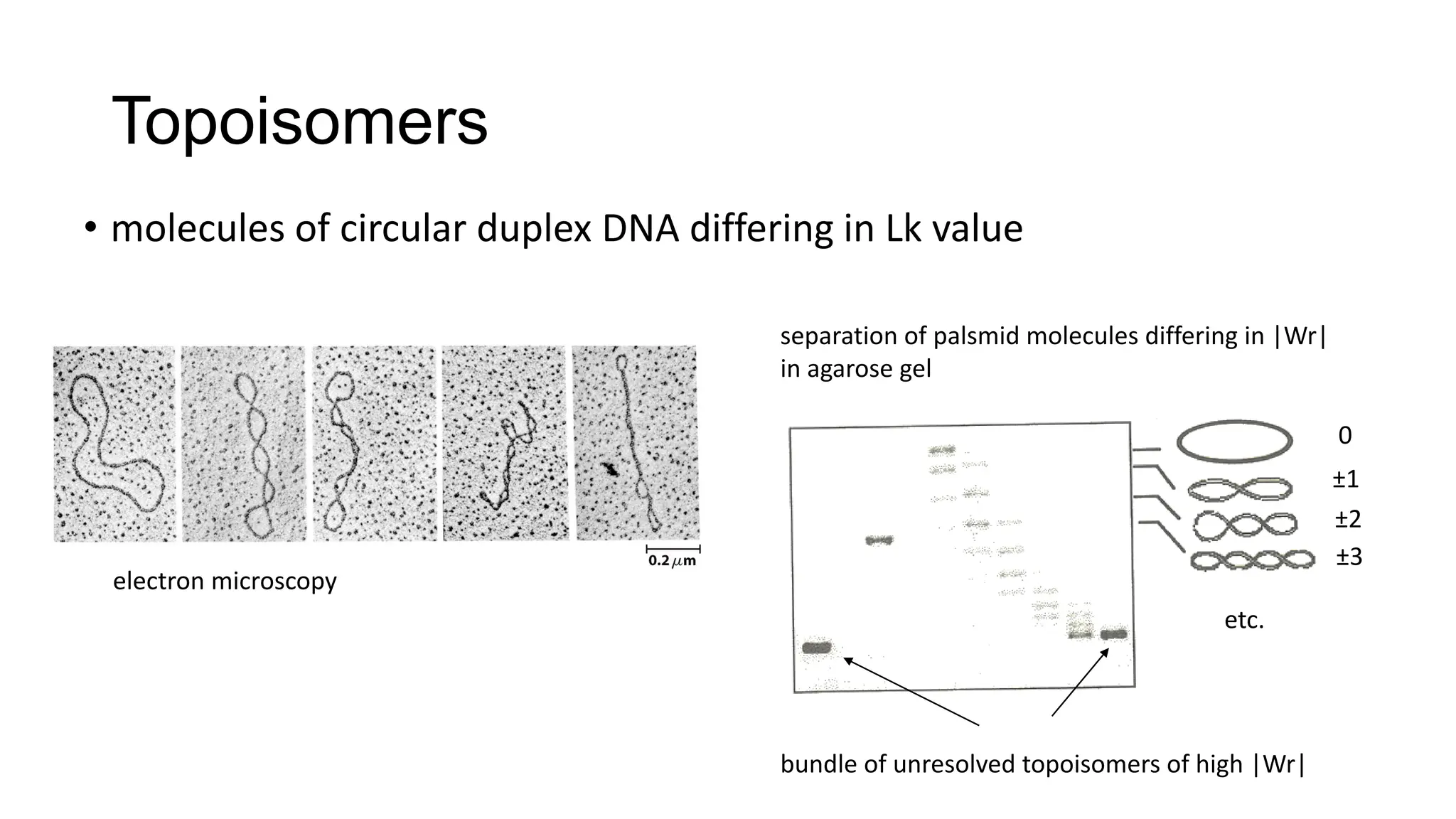
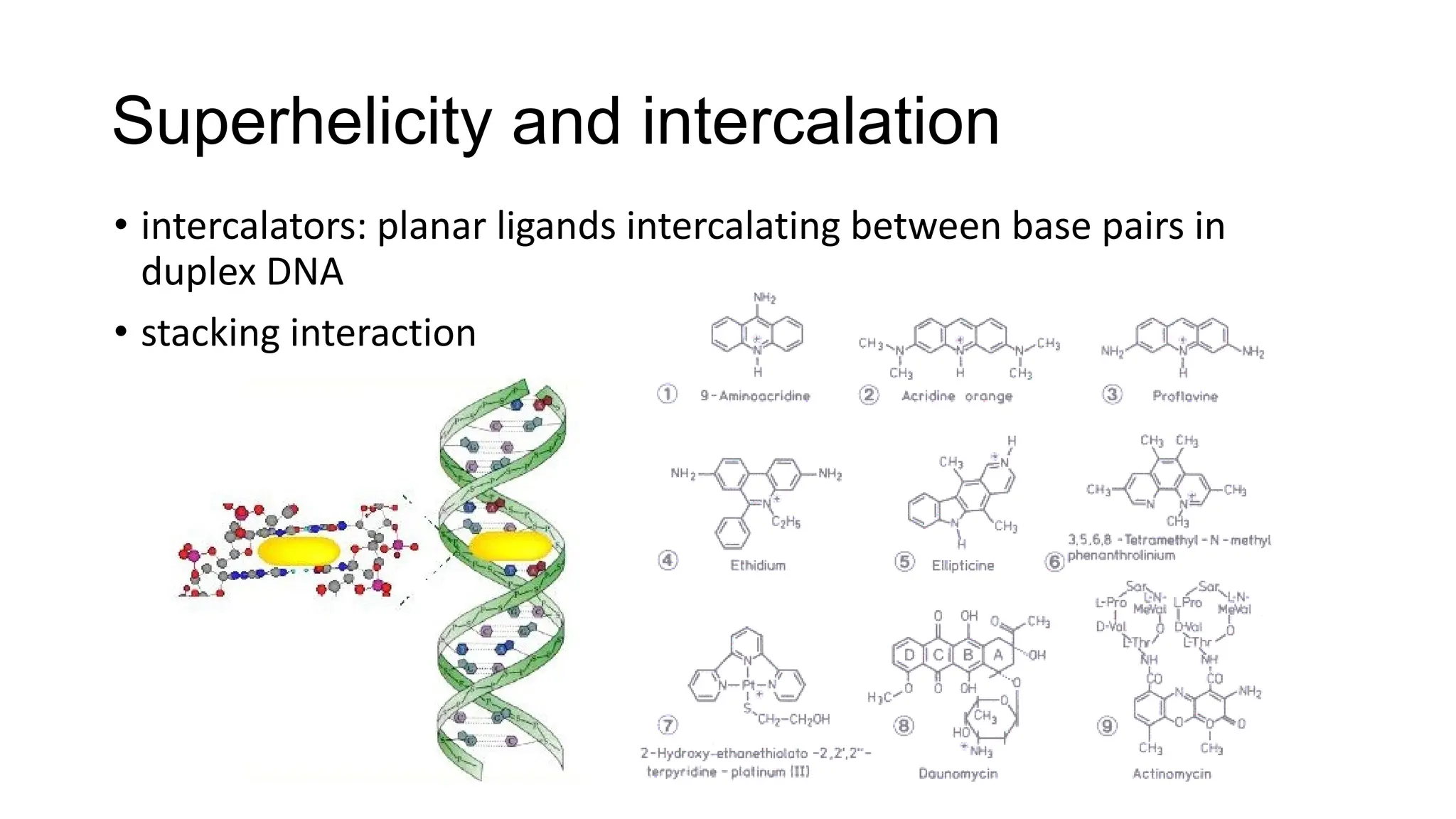
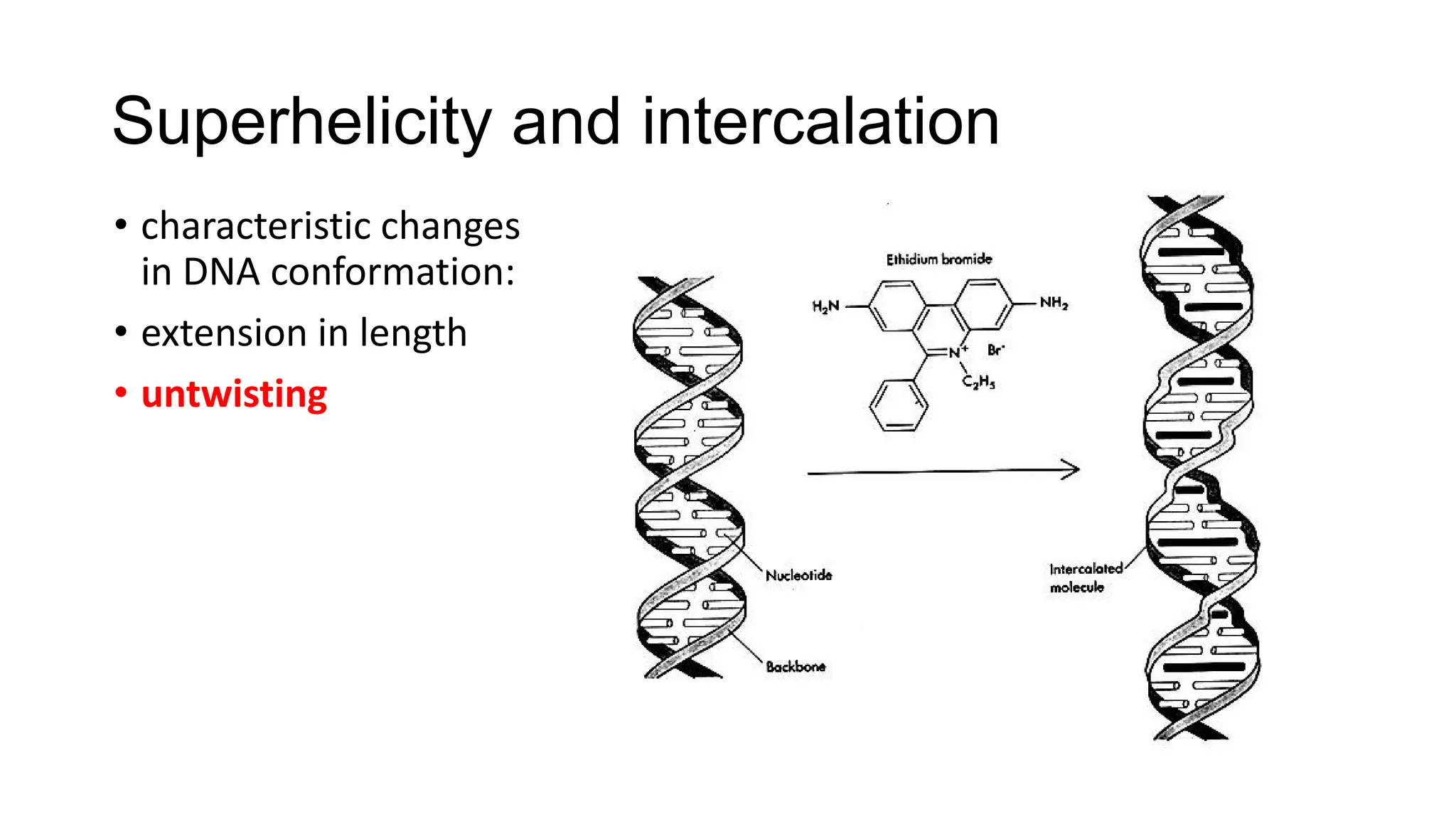
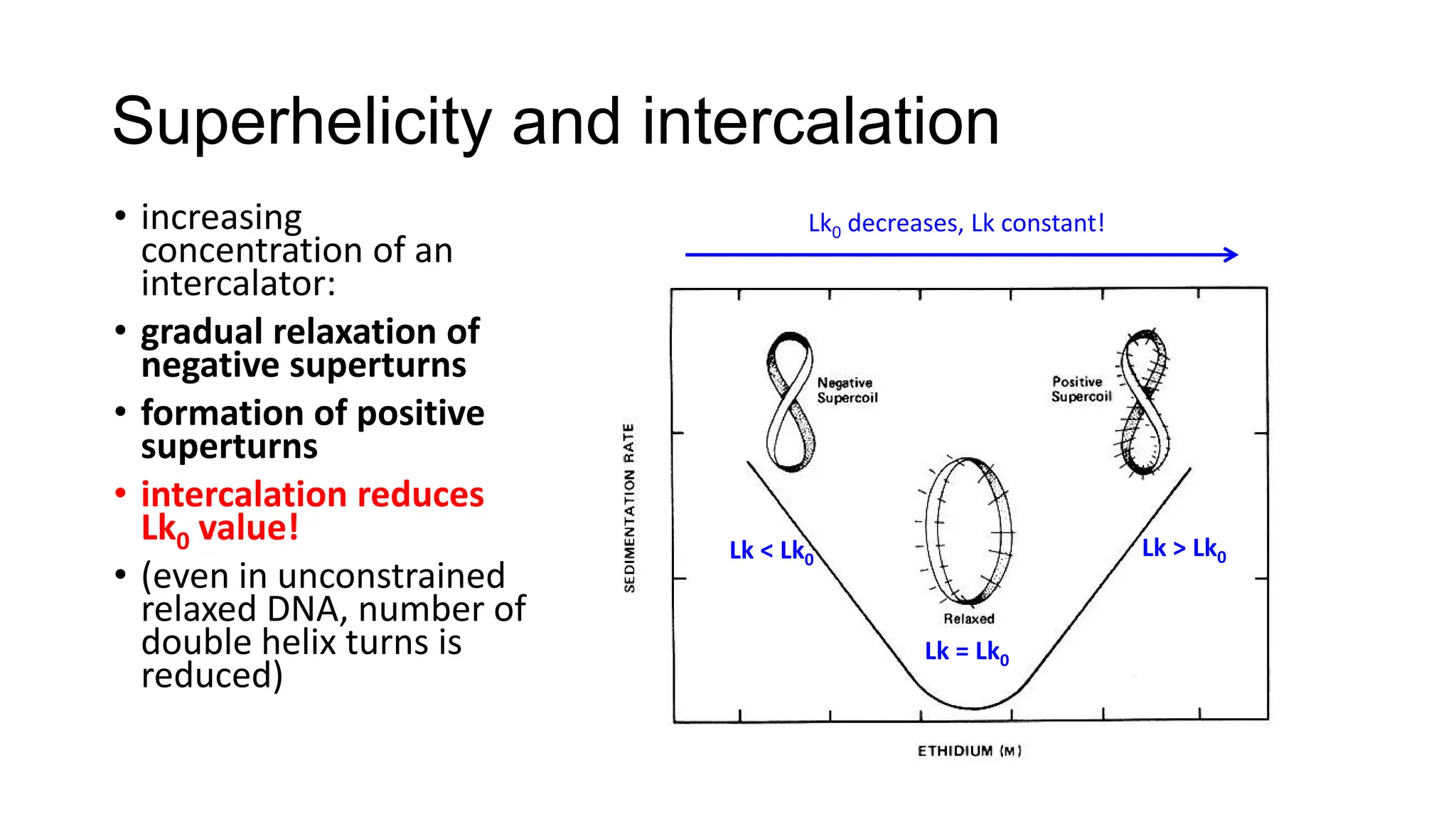
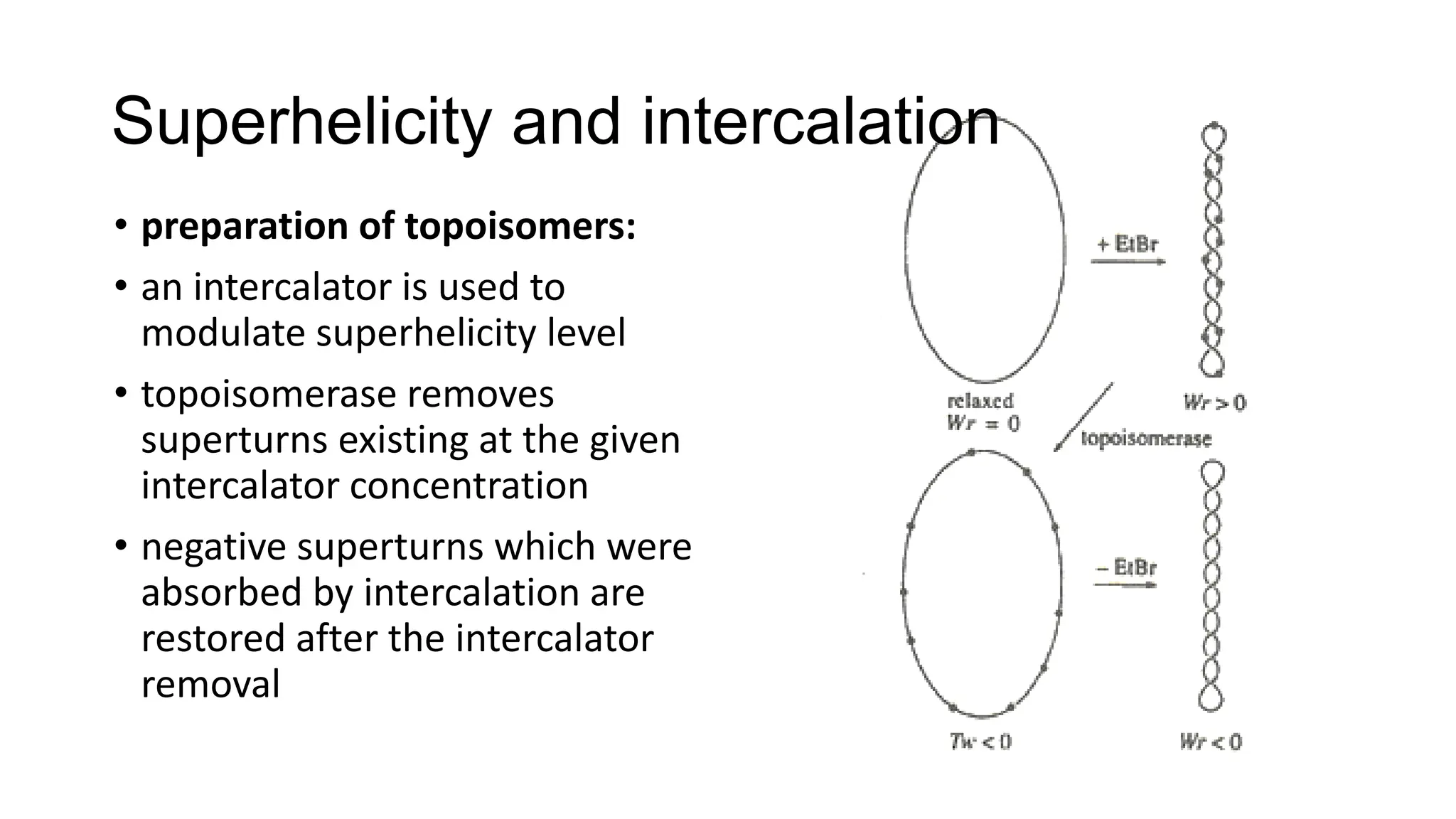
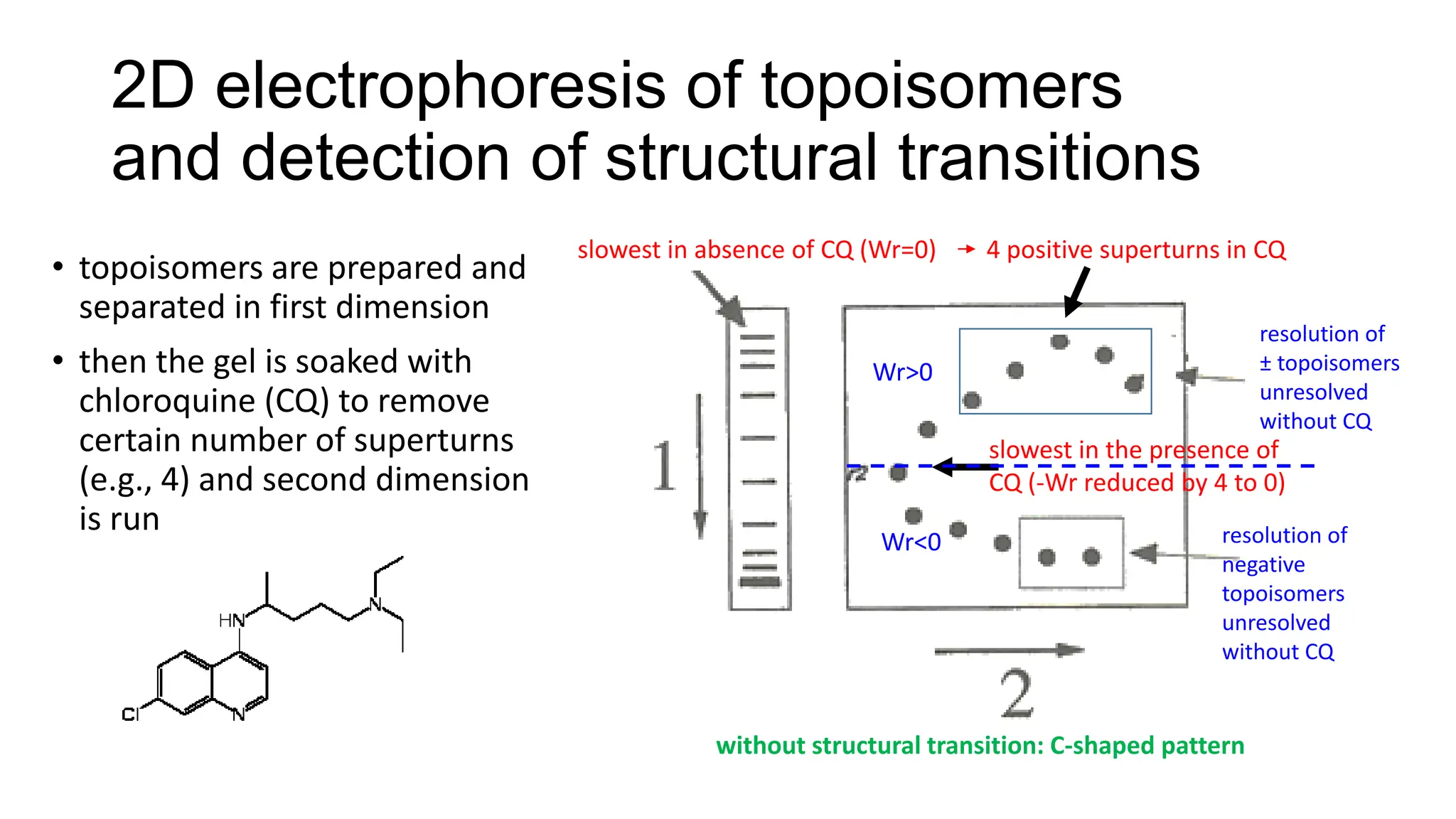
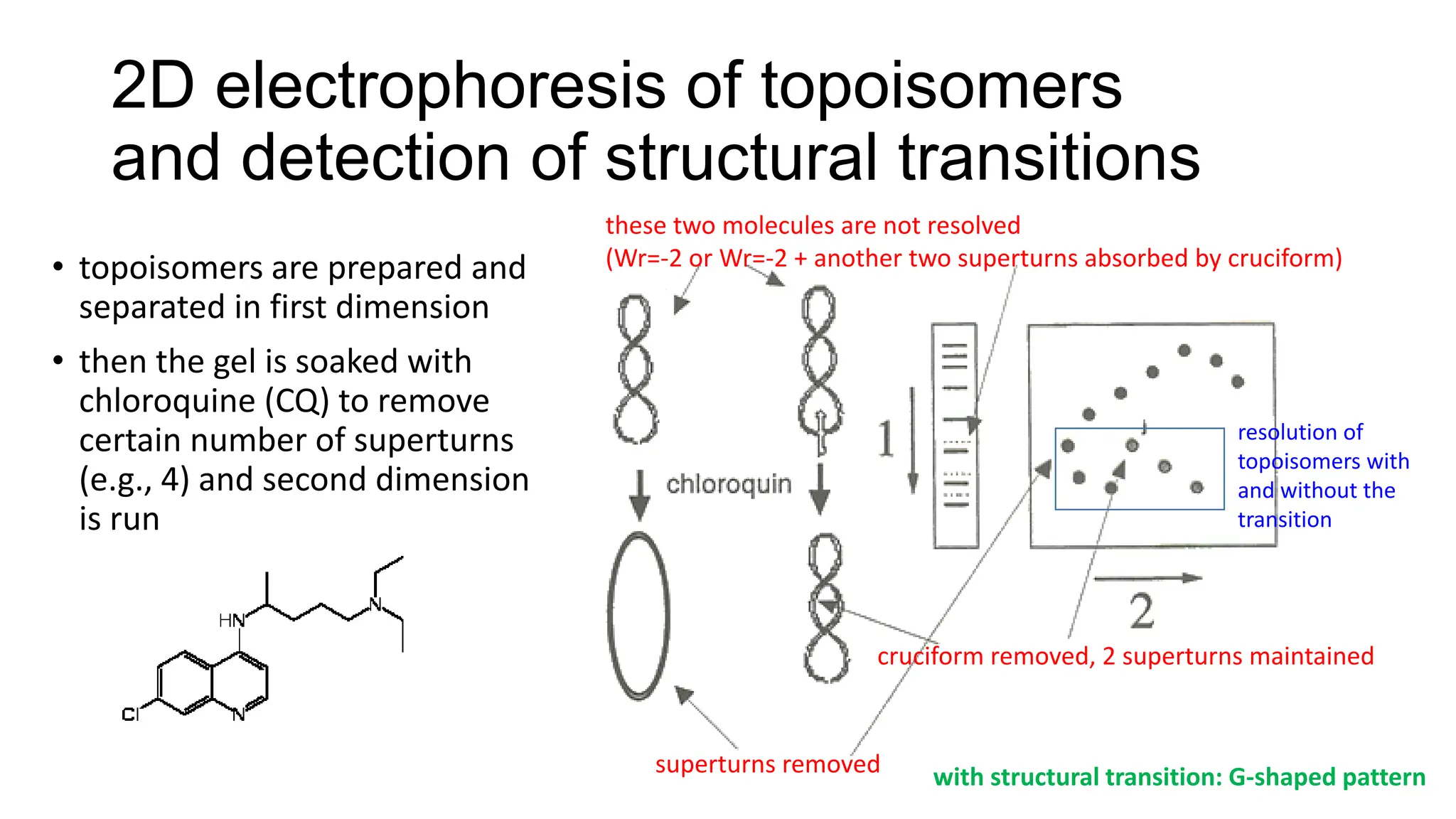
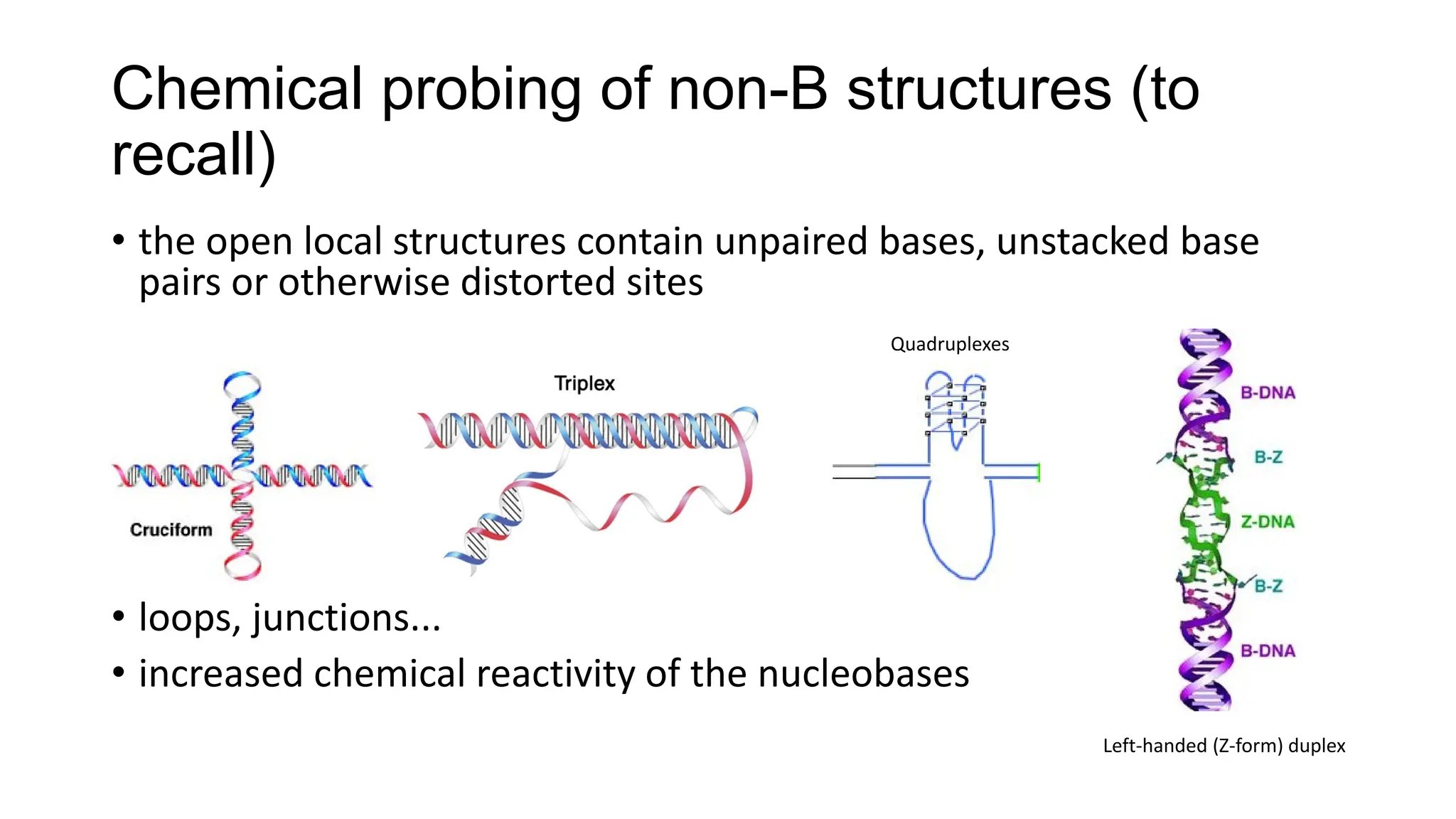
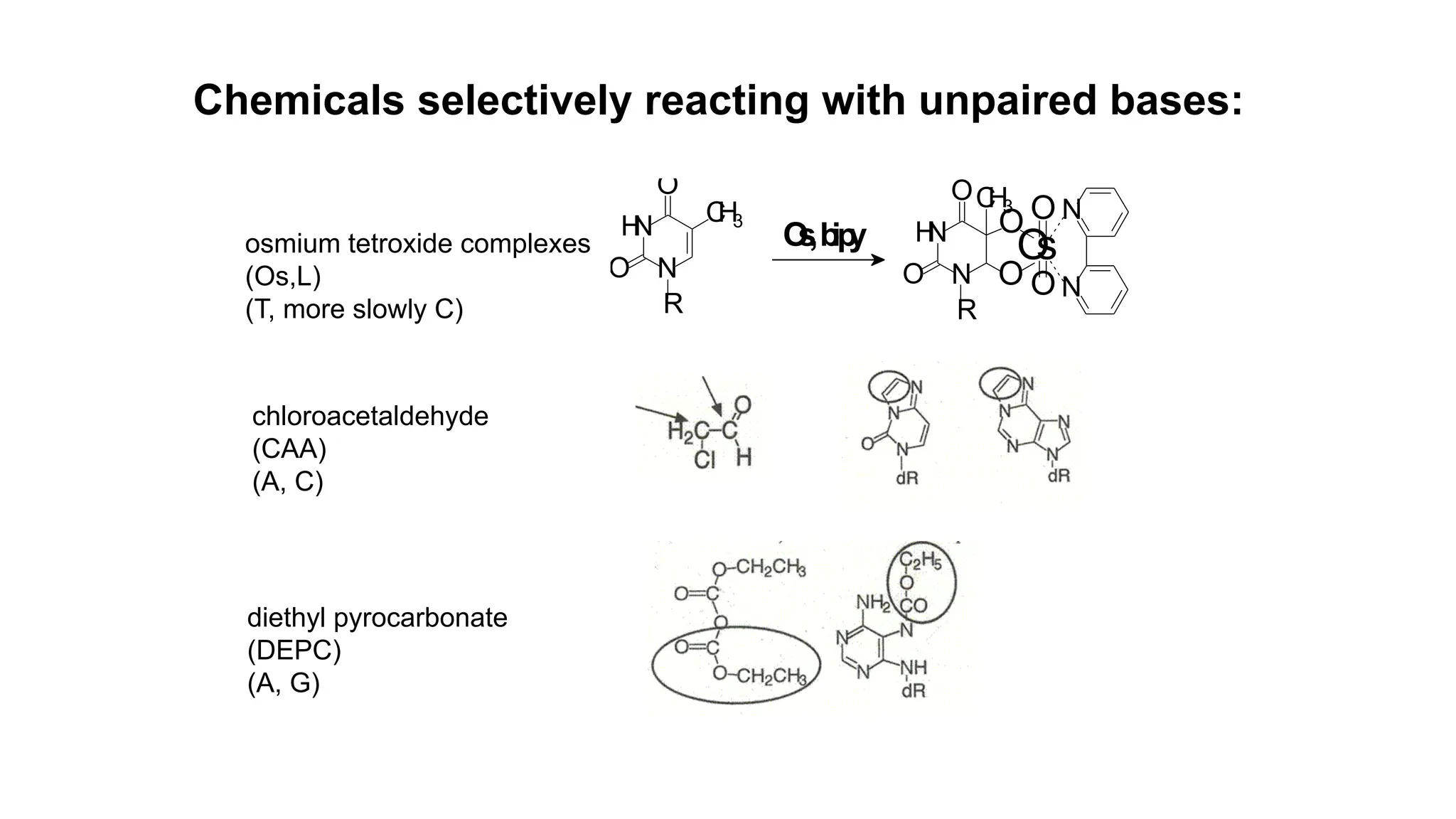
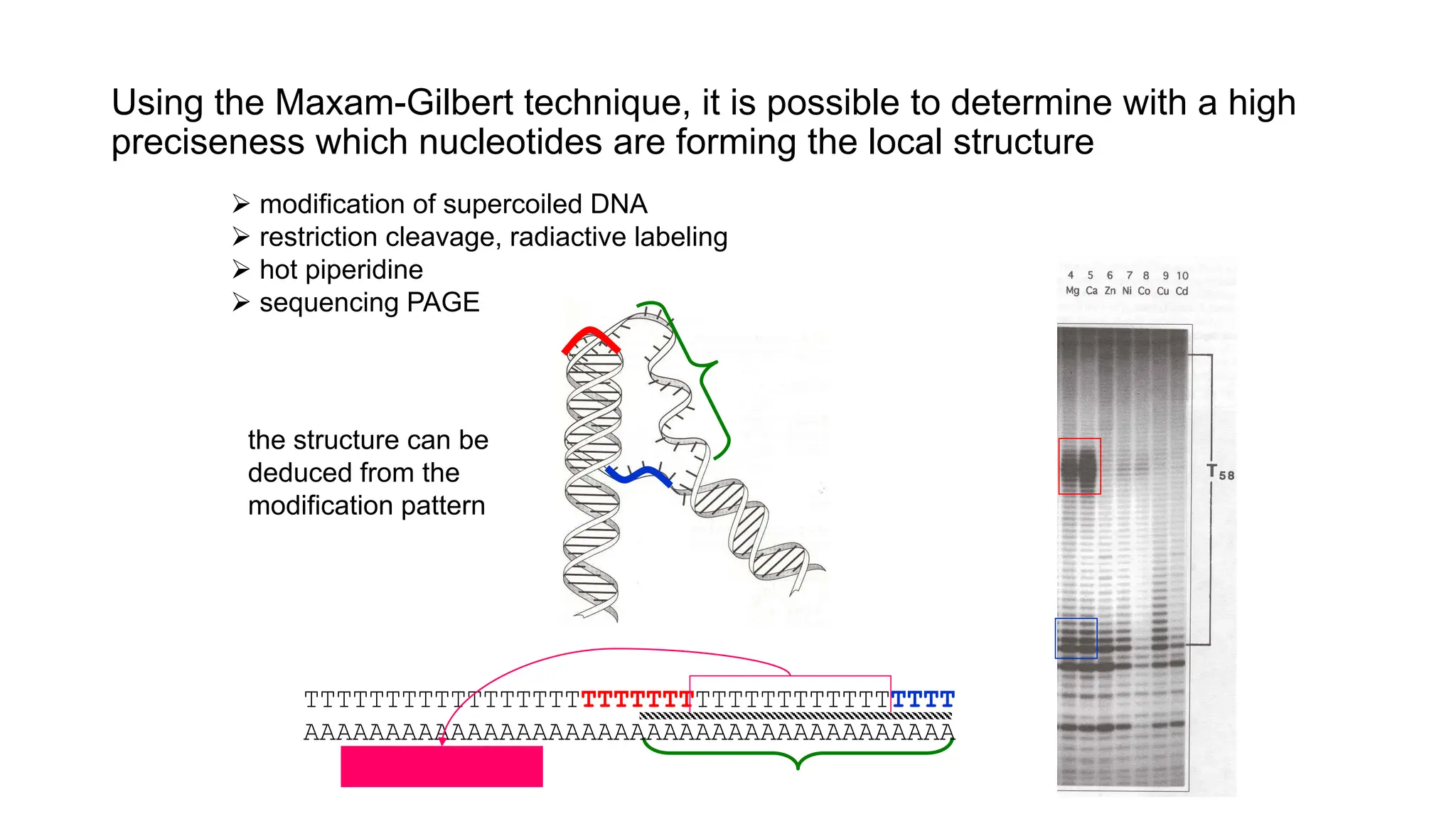
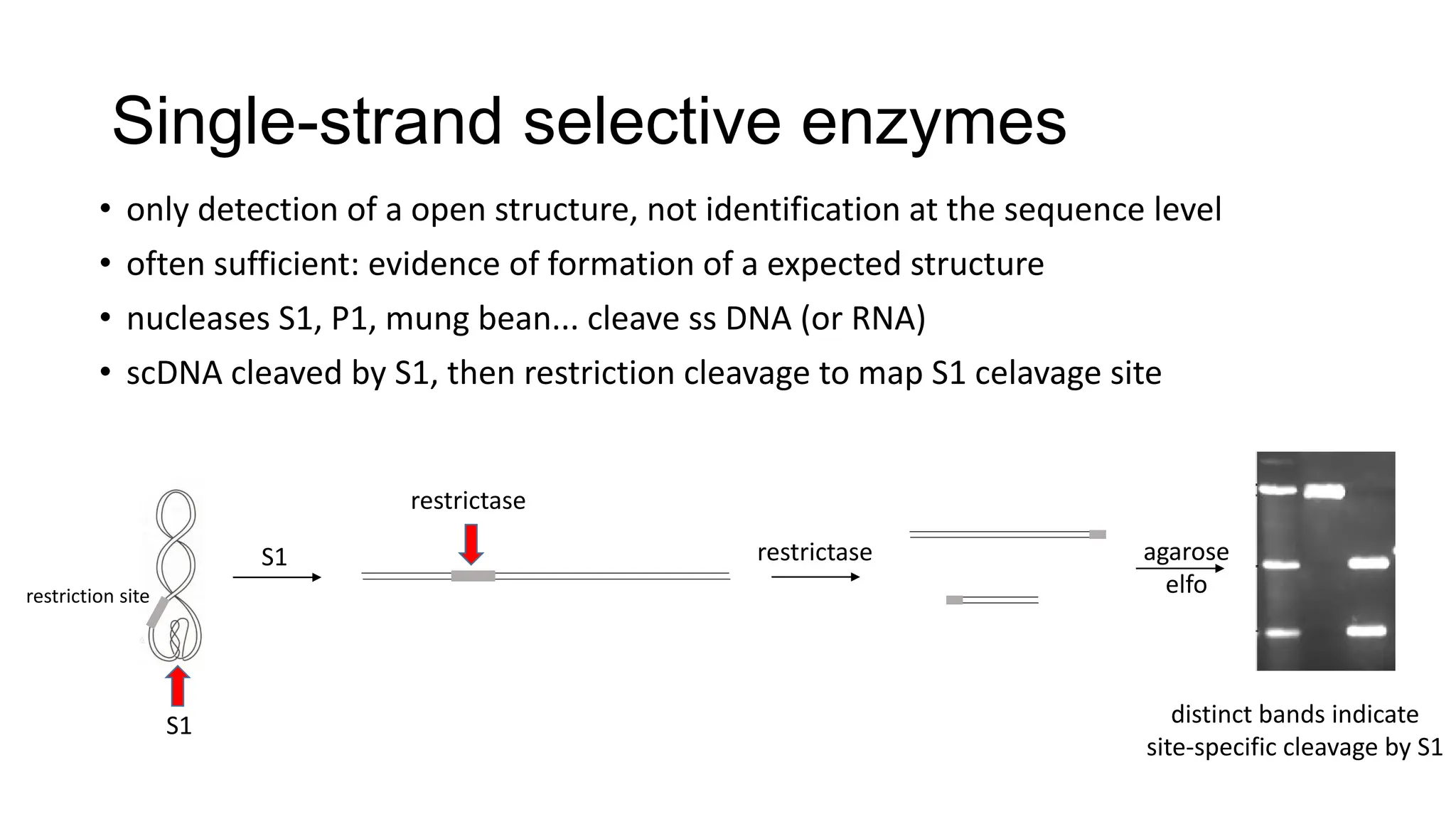
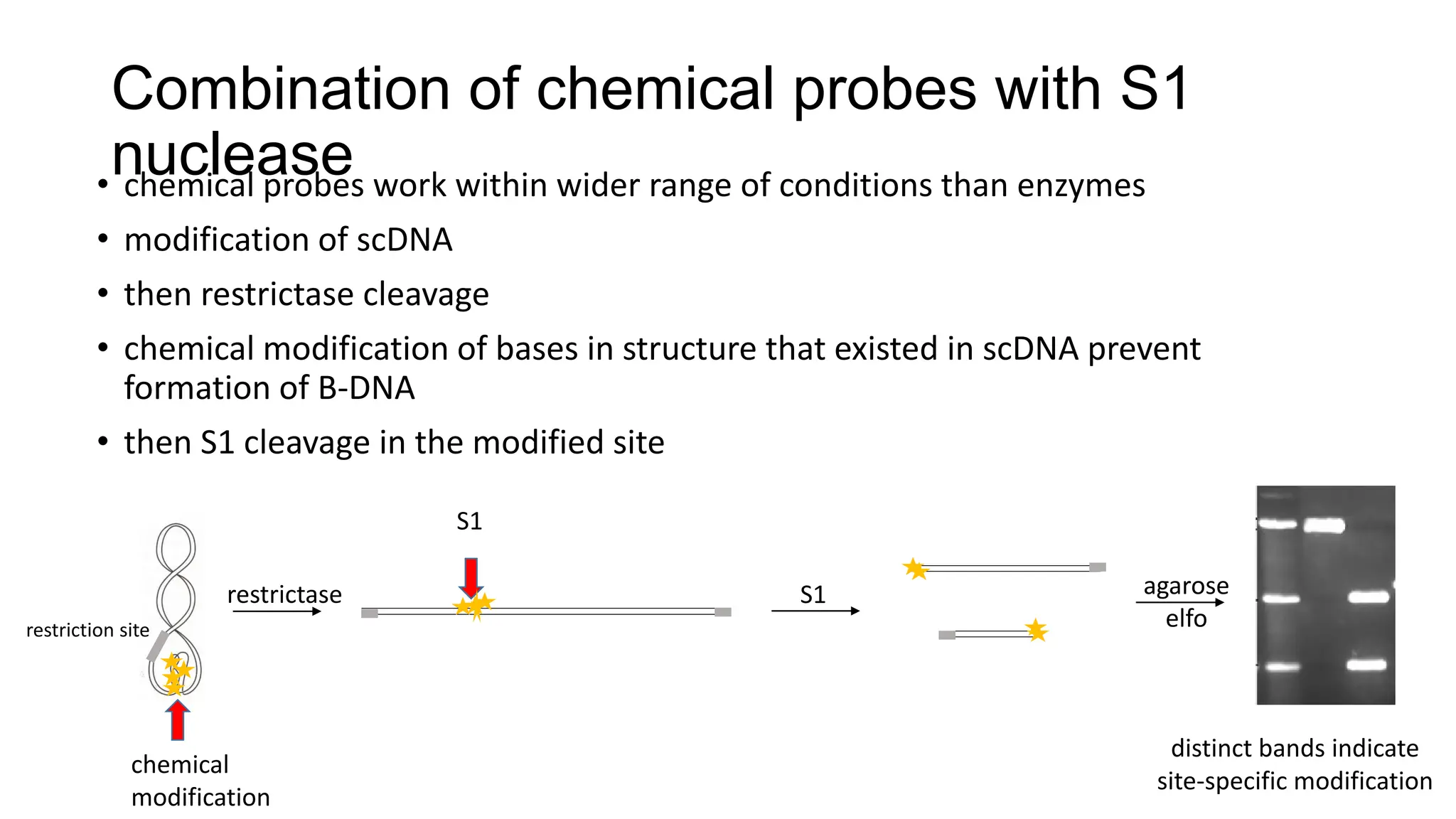
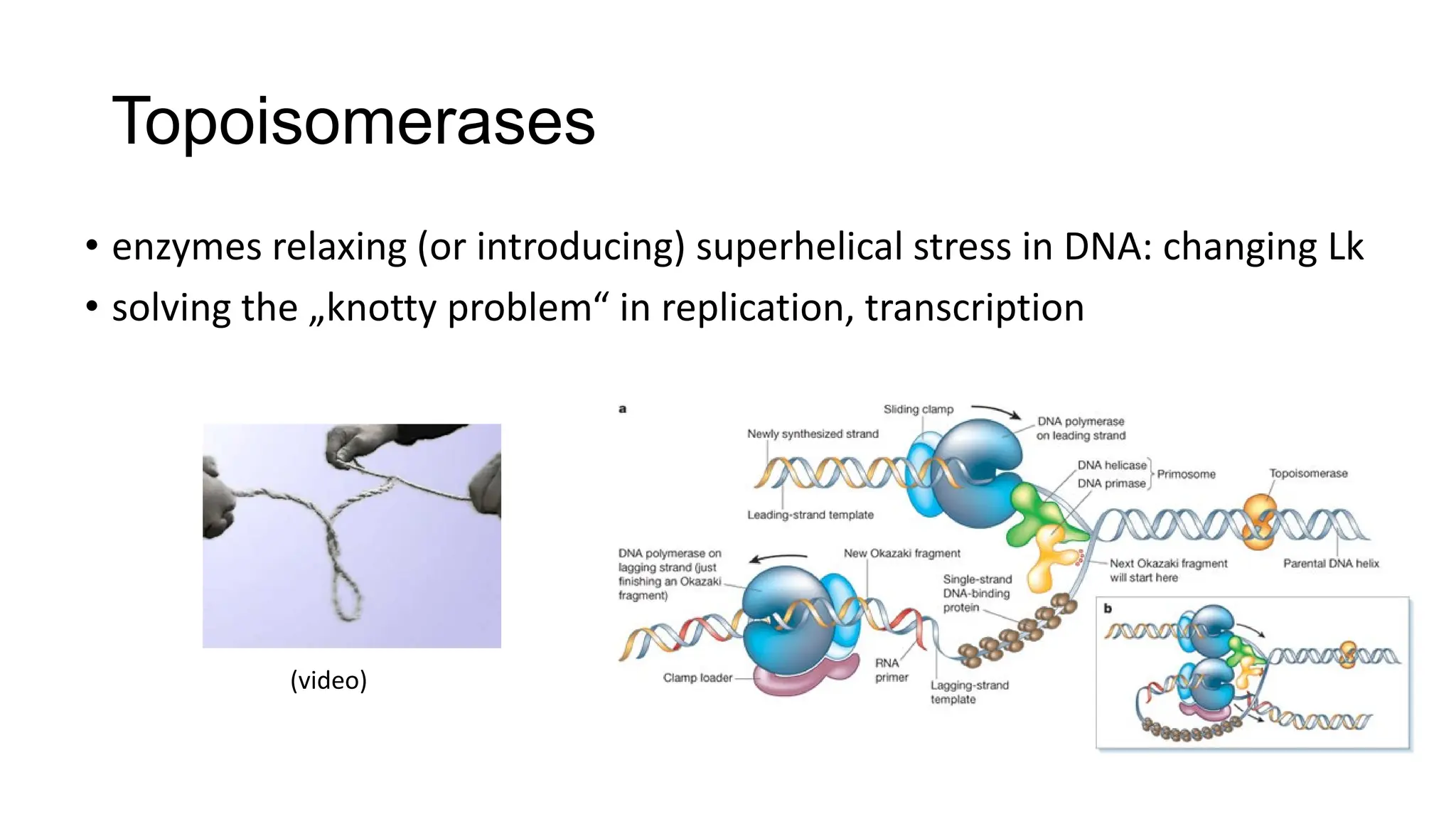
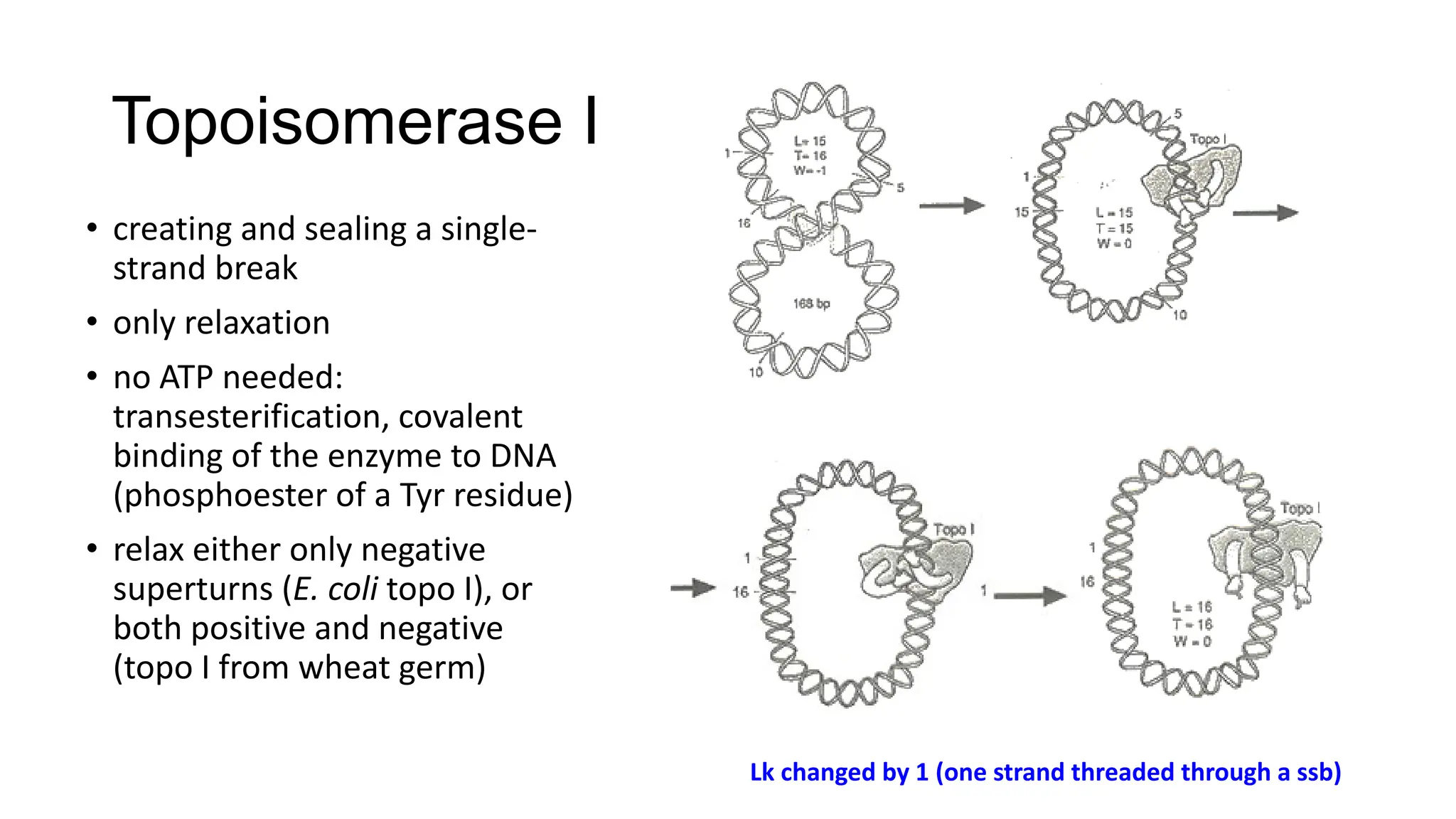
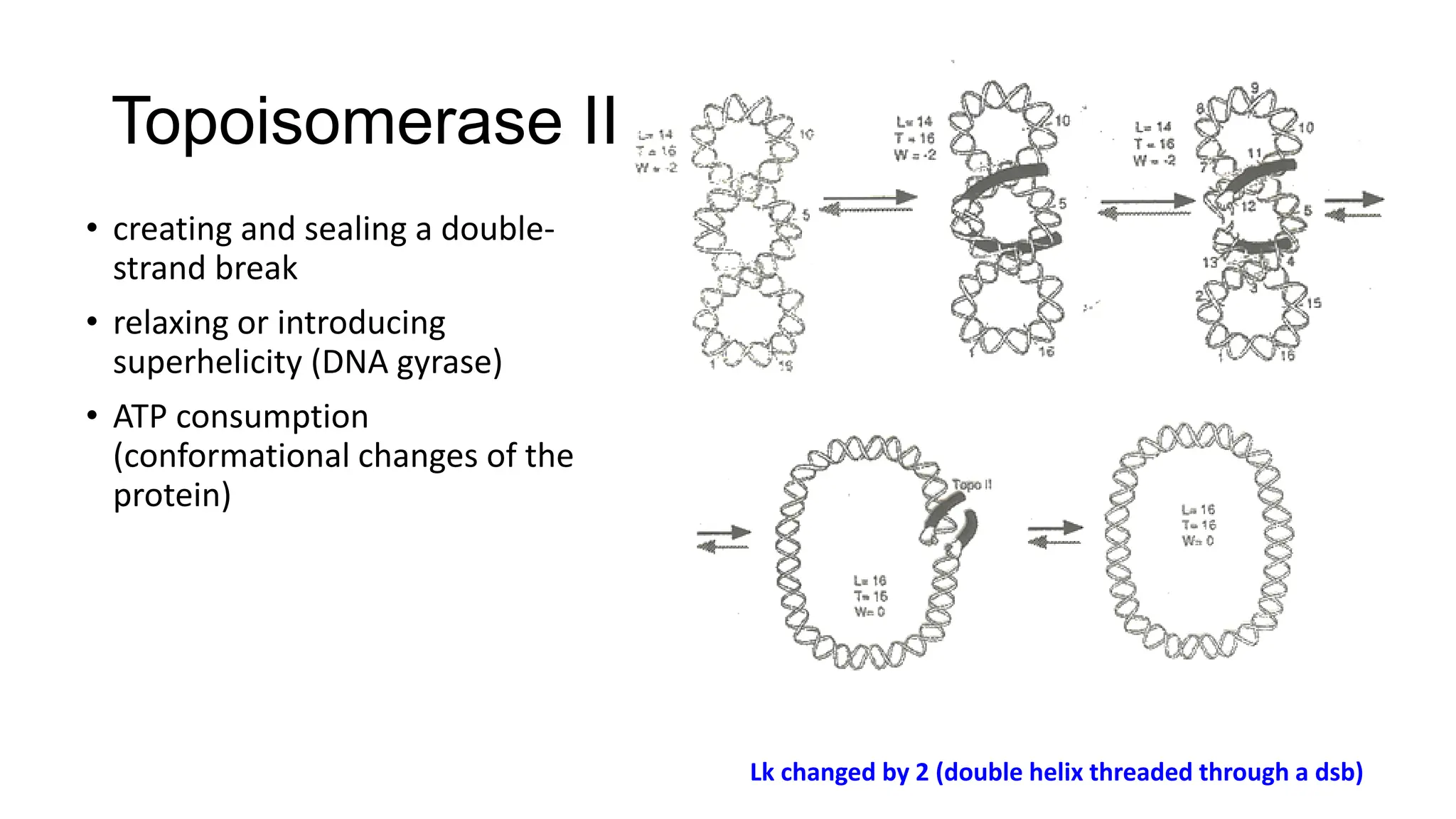
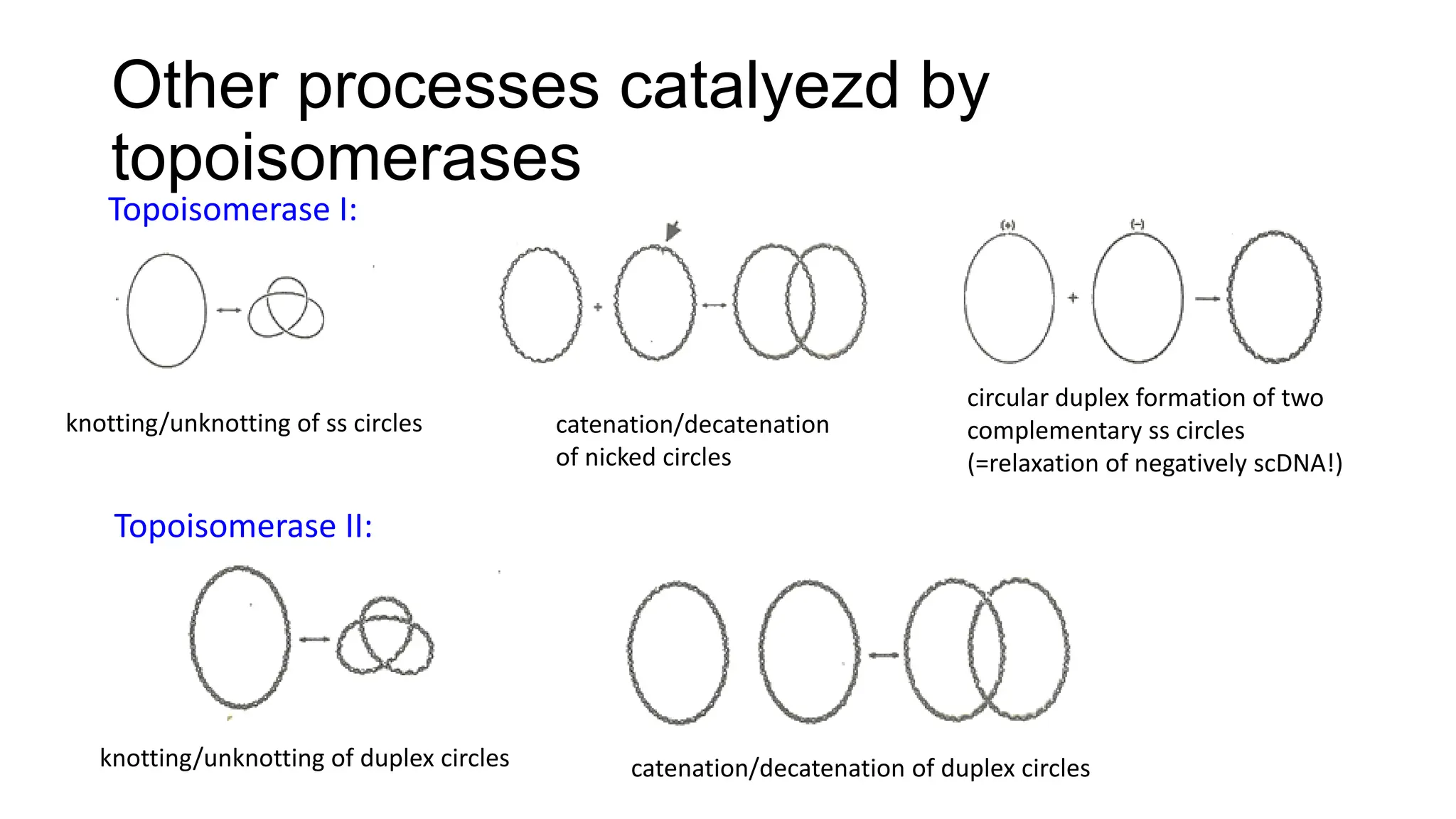
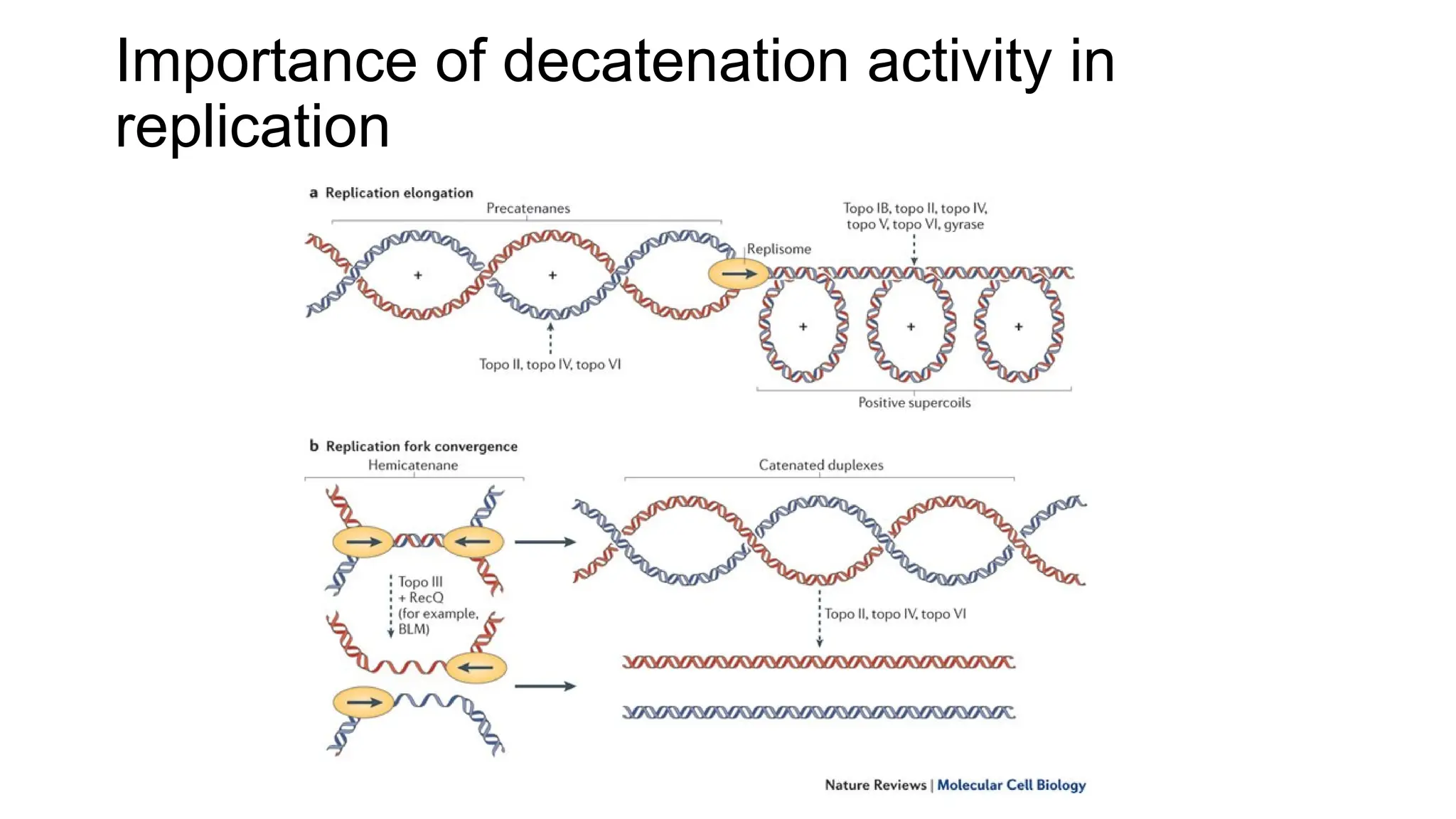
The document discusses DNA supercoiling, highlighting how circular DNA exists in supercoiled and relaxed forms, influencing its elasticity and structural properties. It details the interactions of DNA with proteins and the impact of local untwisting during processes like replication and transcription, as well as the role of topoisomerases in managing superhelical stress. Additionally, various experimental techniques for studying DNA structure and dynamics, including sedimentation and electrophoresis, are outlined.