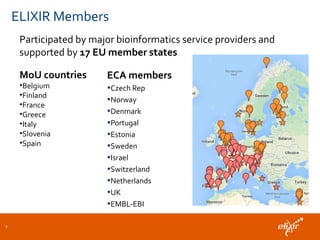
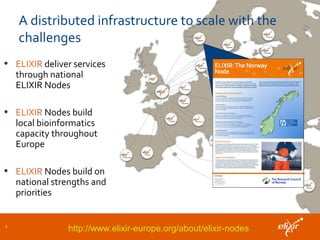
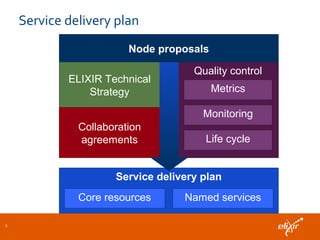
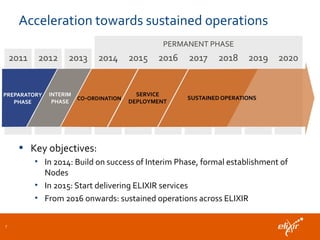
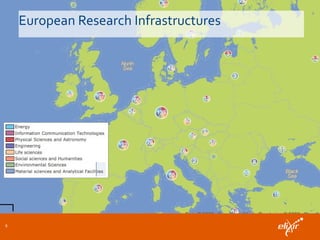
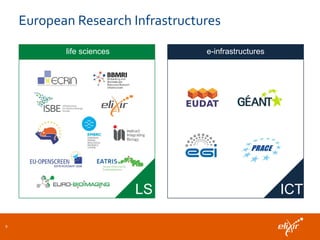
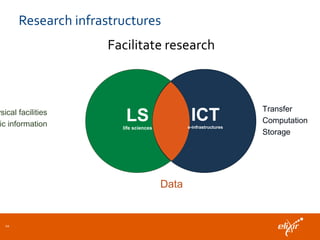
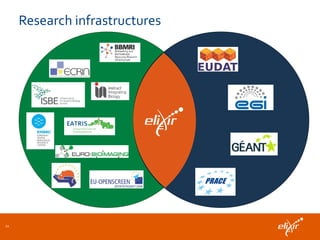
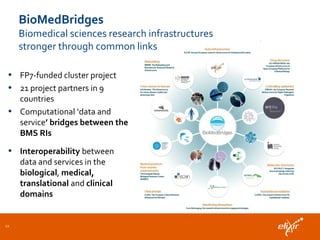
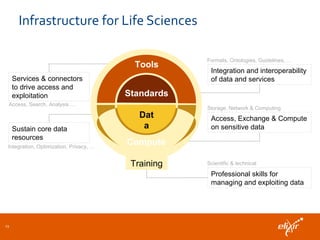
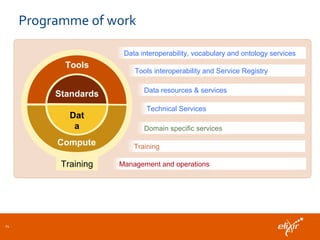
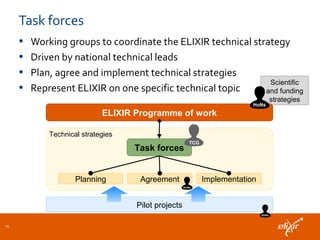
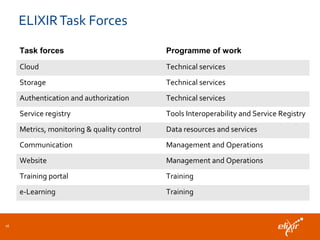
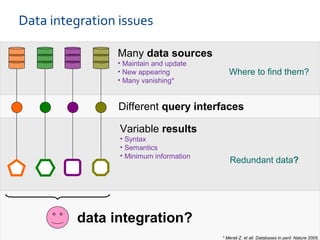
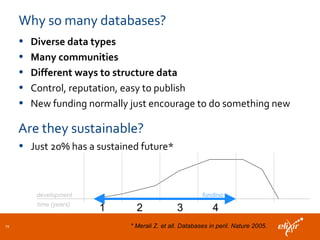
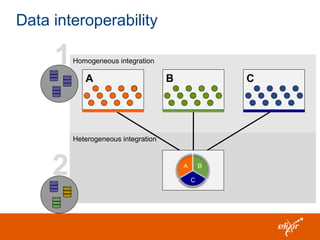
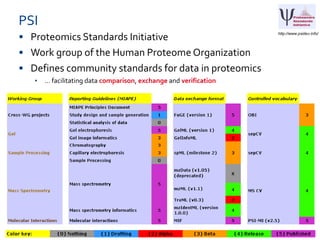
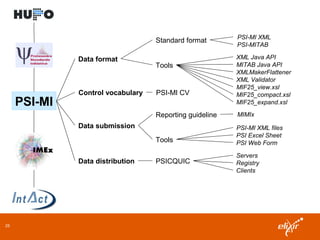
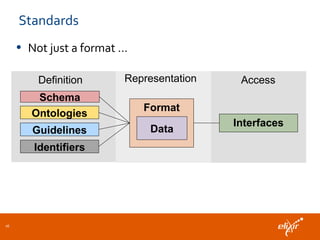
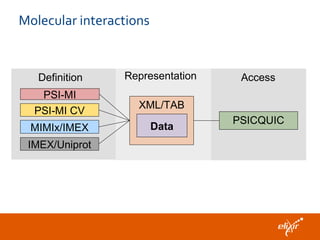
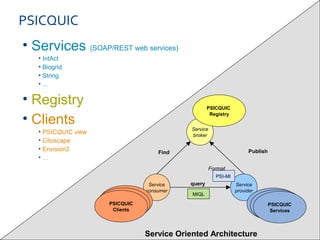
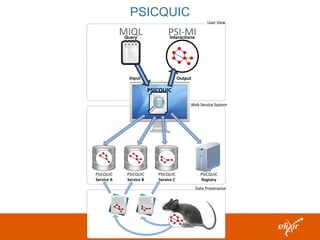
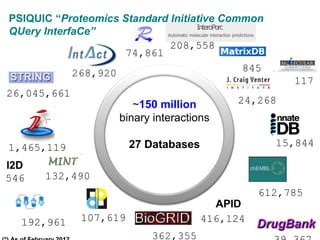
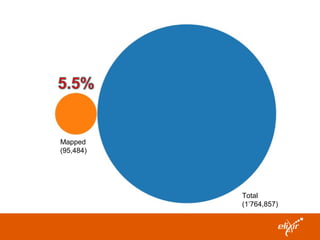
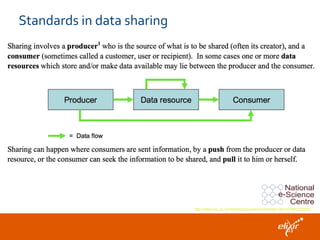
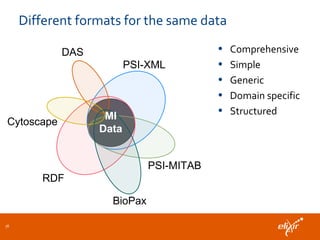
ELIXIR is a European research infrastructure for biological information that aims to facilitate life sciences research across Europe. It brings together life science resources from member countries to build a robust infrastructure for biological data. Individual organizations or countries cannot achieve this alone. ELIXIR establishes national nodes that leverage local strengths and priorities to deliver shared services through a distributed network. This allows the infrastructure to scale effectively with increasing data challenges. ELIXIR also works to improve data integration and interoperability across distributed resources through activities like developing standards and linking related communities.