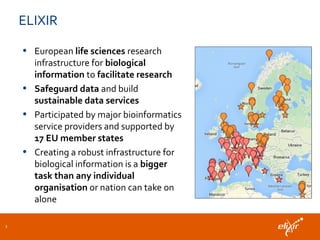
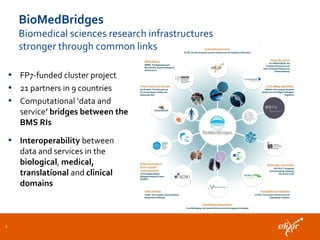
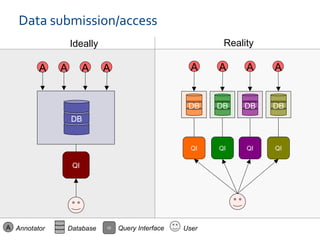
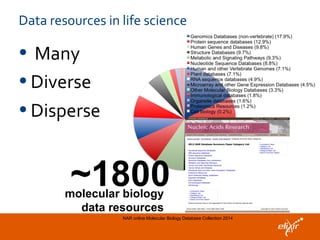
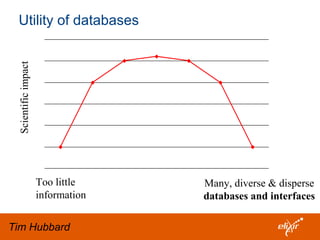
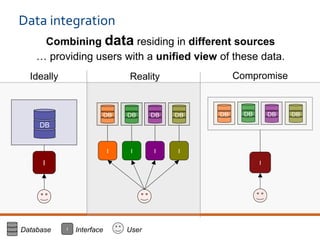
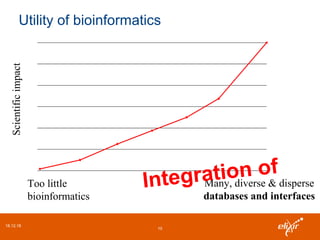
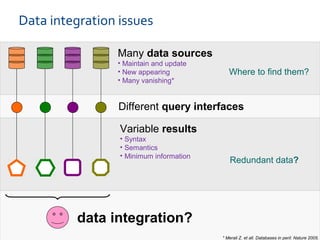
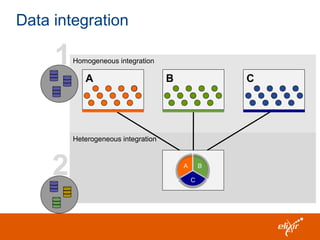
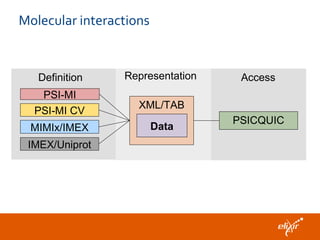
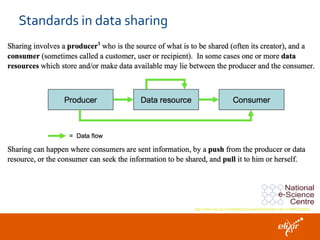
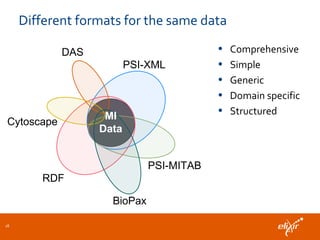
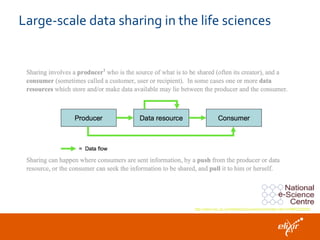
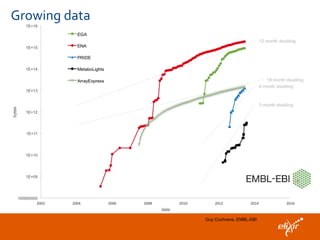
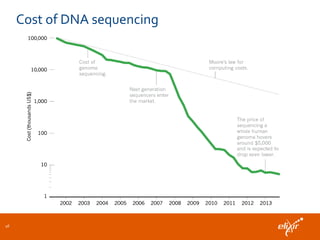
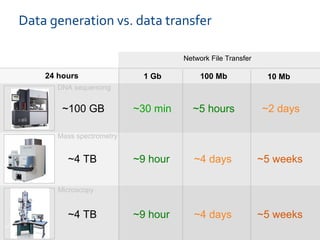
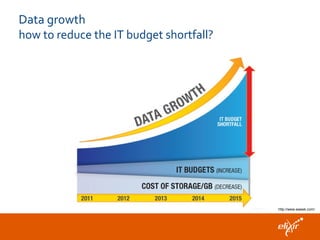
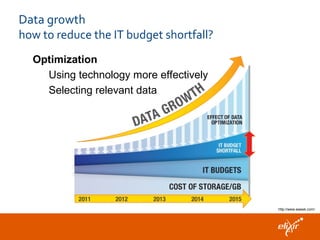
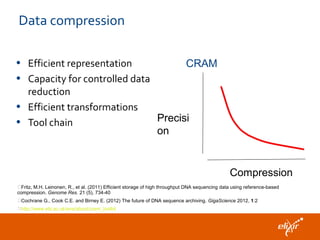
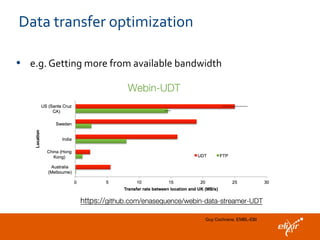
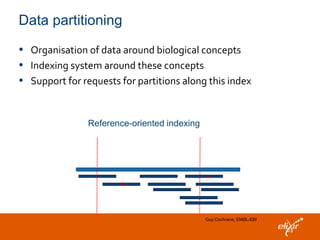
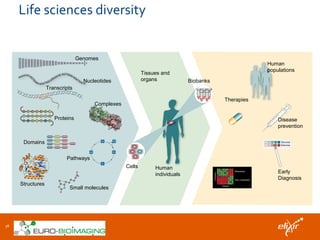
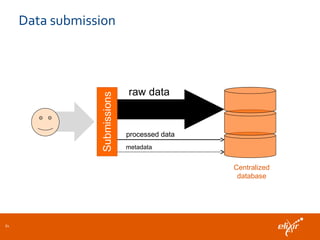
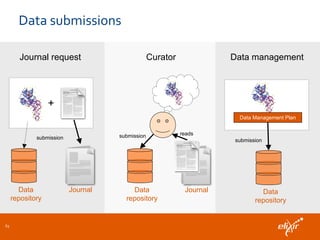
The document discusses the European Life Sciences Infrastructure for Biological Information (ELIXIR) and its role in standardizing and managing big data in the life sciences, emphasizing the importance of interoperability among diverse data sources. It highlights initiatives like Biomedbridges, which connects various biomedical research infrastructures to address societal challenges and promote data sharing. The text also covers best practices in identifier management, data integration, and strategies to optimize e-infrastructures for future data demands in life sciences.