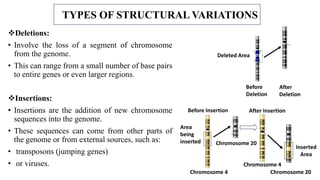
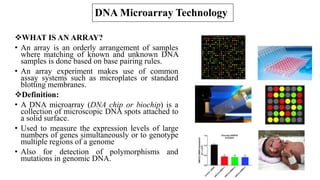
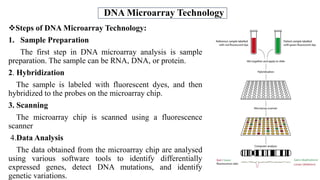
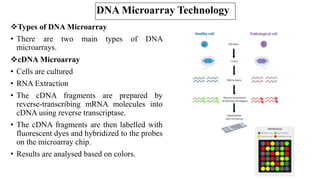
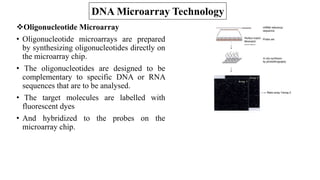
The document discusses structural variations (SVs) in the genome, their types, detection methods, and implications in cancer and other fields. SVs include deletions, duplications, insertions, inversions, and translocations, which significantly contribute to genetic diversity and disease susceptibility. Advanced technologies like DNA microarray and next-generation sequencing have enhanced the detection and analysis of these variations, influencing personalized medicine and raising ethical considerations.