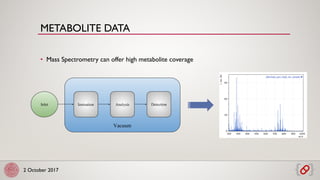
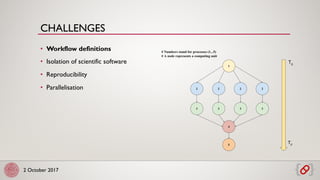
The document discusses scalable and reproducible workflows for biological computations using Pachyderm and Kubernetes. It highlights challenges in data analytics, the importance of containerization for reproducibility, and the features of Pachyderm, including version control for data and pipeline systems. The goal is to enable efficient and effective data processing in biomedicine through container-based workflows.



![2 October 2017
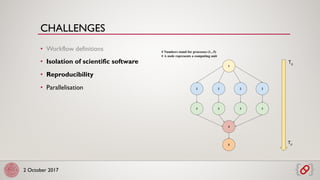
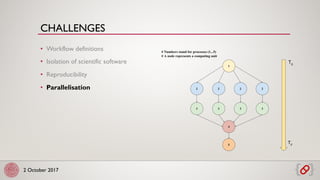
• Deployment, scaling and management of containers in a
cluster
• Kubernetes: big and active community
• Automatic healing and machine decoupling
[1] https://www.kubernetes.io
[1]
CONTAINER ORCHESTRATION TOOLS](https://image.slidesharecdn.com/bdlifesciences-171002113437/85/Scalable-and-reproducible-workflows-with-Pachyderm-8-320.jpg)
![2 October 2017
• Workflow-system based on Kubernetes
• A distributed data processing tool based
on containers
• Enables reproducibility, provenance,
parallelization and isolation
“You can focus on being productive, while
Pachyderm will scale up and analyze for you”
[2] https://www.pachyderm.io
[2]
WHAT IS PACHYDERM?](https://image.slidesharecdn.com/bdlifesciences-171002113437/85/Scalable-and-reproducible-workflows-with-Pachyderm-10-320.jpg)
![2 October 2017
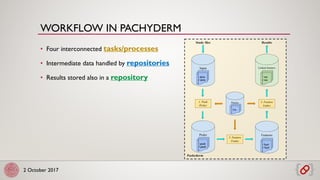
The main primitives are:
• Repositories: versioned collections of data
• Commits: new data
• Files: data storage primitives
[3] https://www.pachyderm.io/pfs.html
[3]
PFS offers version control for data:
PACHYDERM FILE SYSTEM (PFS)](https://image.slidesharecdn.com/bdlifesciences-171002113437/85/Scalable-and-reproducible-workflows-with-Pachyderm-11-320.jpg)
![2 October 2017
• Tasks executed by Kubernetes pods
• Parallelization: spreading data
• Incrementality and glob patterns
• Directed Acyclic Graph
[4] https://www.pachyderm.io/pps.html
[4]
PACHYDERM PIPELINE SYSTEM (PPS)](https://image.slidesharecdn.com/bdlifesciences-171002113437/85/Scalable-and-reproducible-workflows-with-Pachyderm-12-320.jpg)