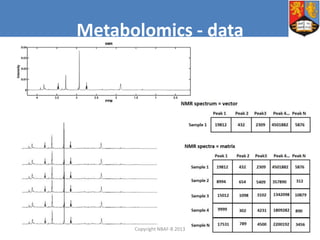
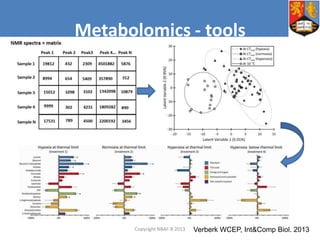
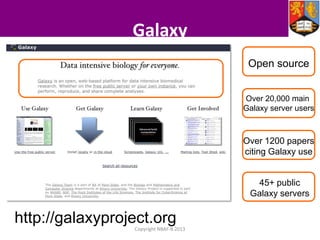
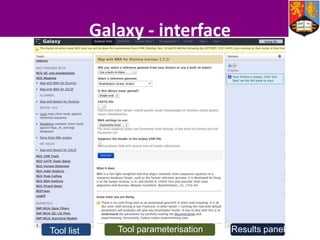
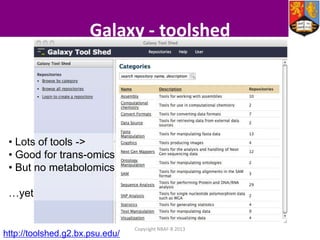
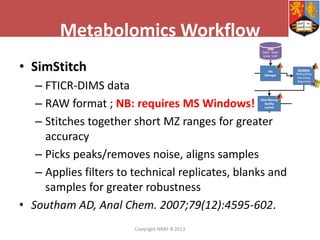
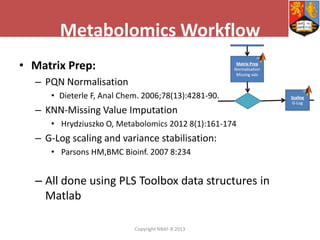
This document describes how Galaxy can be used to analyze metabolomics data. It outlines a metabolomics workflow developed for Galaxy that incorporates tools for processing raw mass spectrometry data files, identifying peaks, normalizing data, and performing multivariate and univariate statistical analyses. The workflow allows metabolomics data to be analyzed within Galaxy while taking advantage of existing computing resources like desktop licenses and processors through Galaxy's Light Weight Runner. This provides the benefits of Galaxy for reproducibility and sharing workflows without needing to transfer large data files or require additional servers.