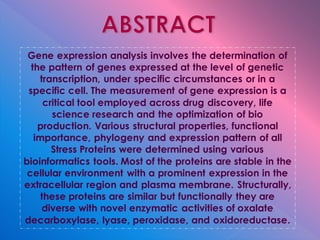
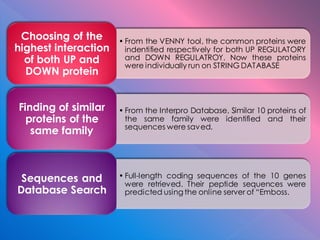
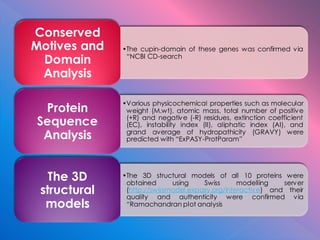
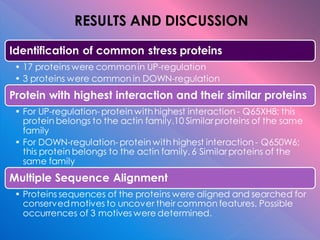
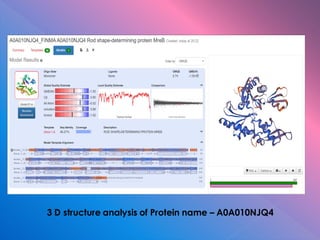
The document summarizes research on stress response proteins in rice (Oryza sativa). It identifies 17 proteins commonly up-regulated and 3 commonly down-regulated in response to drought, heat, and salinity stress. It analyzes the protein with the highest interaction for each group and identifies 10 similar proteins in each family. It examines the proteins' physicochemical properties, 3D structures, and functions in plant defense. The study finds the proteins structurally similar but functionally diverse, concluding they help rice cope with stress through complex regulatory interactions.