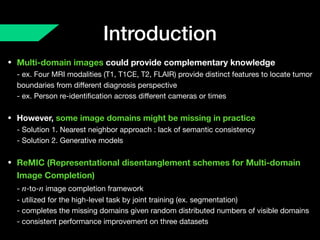
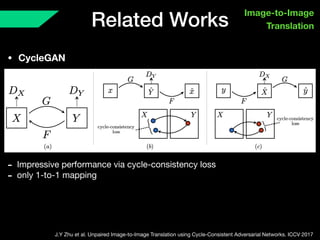
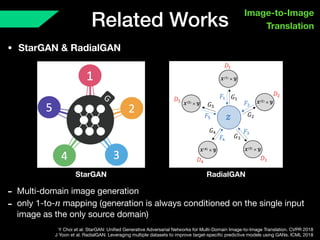
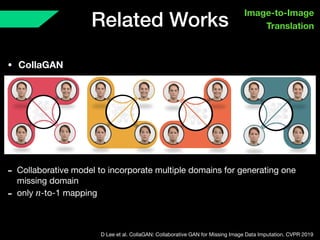
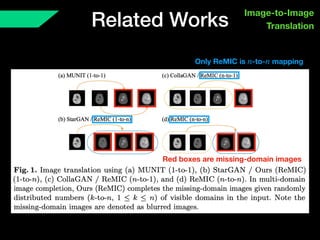
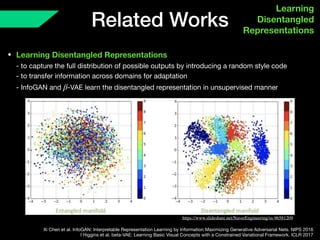
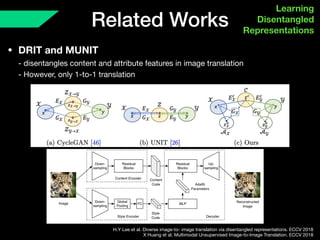
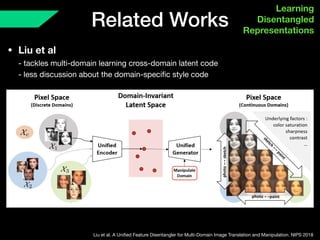
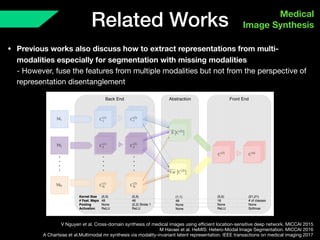
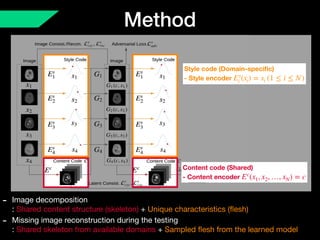
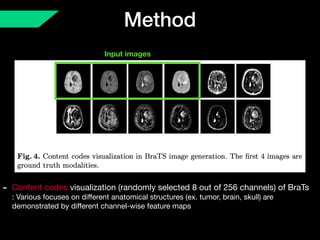
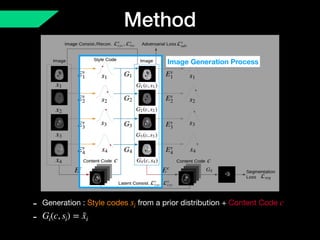
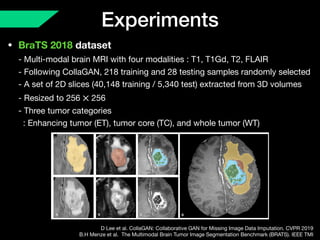
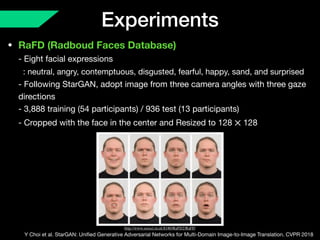
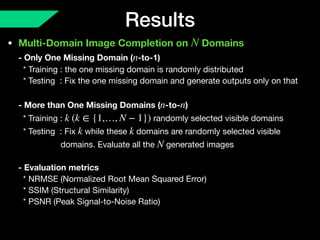
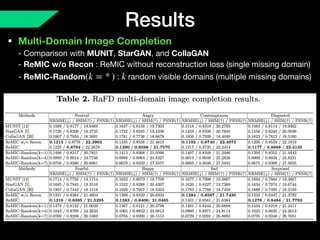
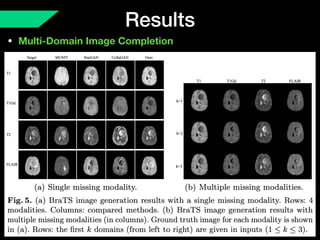
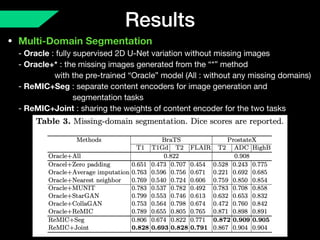
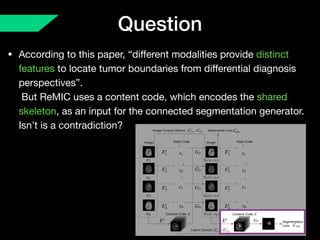
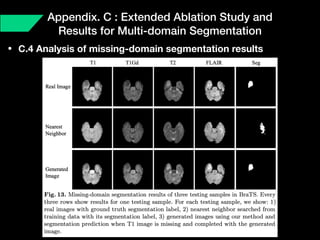
The document presents a framework named REMIC for multi-domain image completion, addressing challenges when certain image domains are missing. It integrates shared content encoding and domain-specific style encoding to complete missing information effectively across different datasets, demonstrating significant performance improvements. The framework is applicable to both medical and natural images and is evaluated on various datasets with detailed results compared to existing models.