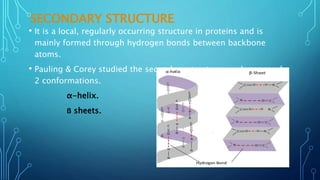
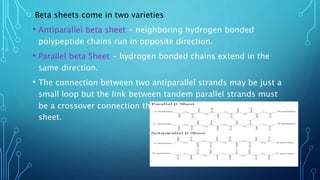
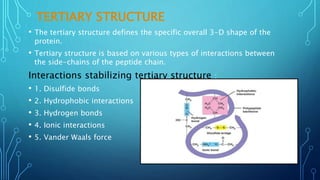
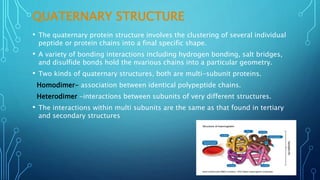
This document provides an overview of protein structure and function. It discusses the four levels of protein structure: primary, secondary, tertiary, and quaternary structure. The primary structure is the amino acid sequence. Secondary structures include alpha helices and beta sheets formed by hydrogen bonding. Tertiary structure describes the overall 3D shape formed by interactions between amino acid side chains. Quaternary structure involves the clustering of multiple peptide chains. Finally, it outlines several key functions of proteins, including structural proteins, transport proteins, and enzymes/receptors.


![AMINO ACID
• Amino acids are a group of organic compounds containing two
functional groups – amino and carboxyl.
• The amino group [ -NH₂] is basic while the carboxyl group [- COOH] is
acidic in nature.
• There are about 300 amino acids occur in nature. Only 20 of them occur
in proteins.](https://image.slidesharecdn.com/proteinstructureandfunctionpptmdmobarakhossain-230910174457-83f5e4c3/85/PROTEIN-STRUCTURE-AND-FUNCTION-PPT-MD-MOBARAK-HOSSAIN-pptx-4-320.jpg)