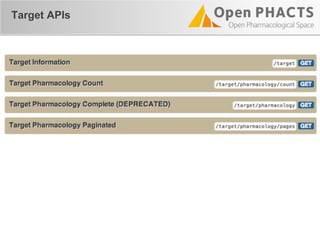
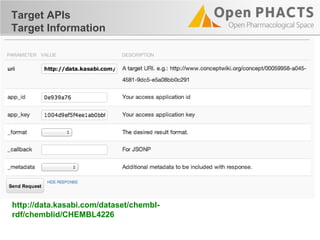
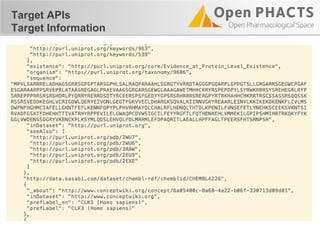
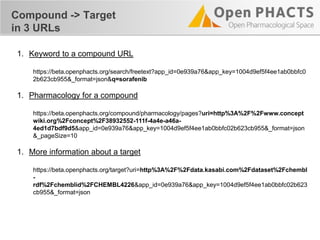
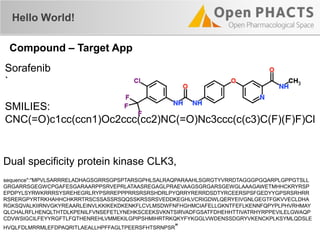
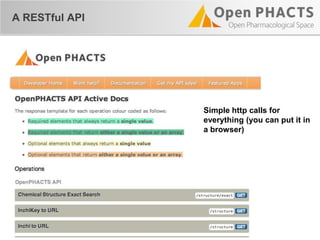
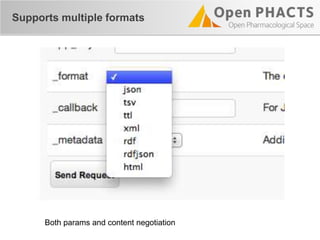
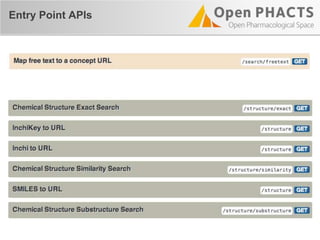
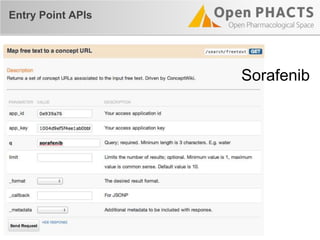
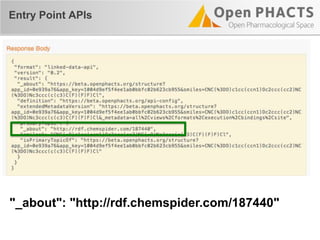
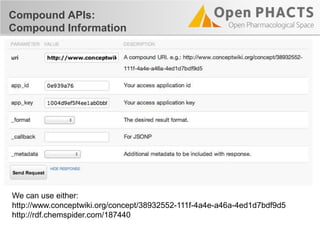
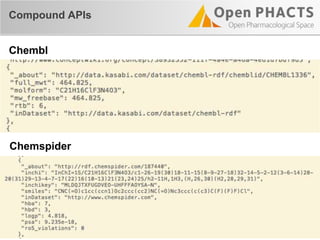
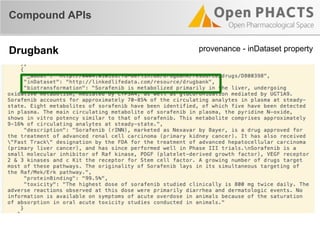
The document outlines the Open PHaCTS (Pharmacology and Chemistry System) API, detailing its capabilities to integrate and access a wide range of pharmaceutical data from various databases. It emphasizes features such as strong chemistry representation, community curation tools, and a developer-friendly interface, which allows users to query interconnected datasets seamlessly. Additionally, it highlights the support for multiple programming languages and active development, with a focus on ensuring easy data retrieval through simple API calls.




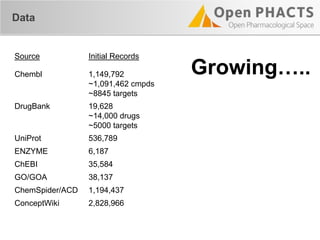




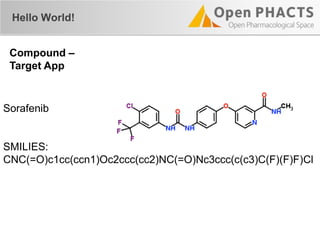





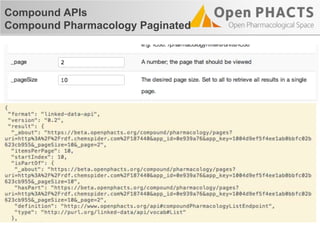
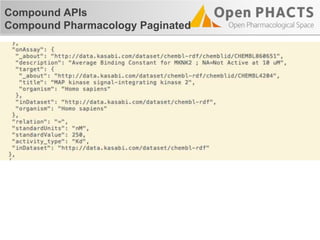
![Compound APIs
Compound Pharmacology Paginated
"items": [
{
"_about": "http://data.kasabi.com/dataset/chembl-rdf/activity/a1650069",
"pmid": "15711537",
"forMolecule": {…},
"onAssay": {
…
"target": {
"_about": "http://data.kasabi.com/dataset/chembl-rdf/chemblid/CHEMBL4226",
"title": "Dual specificity protein kinase CLK3",
"organism": "Homo sapiens"
},
…
},
…
]](https://image.slidesharecdn.com/ops-developer-webinar-july312013-130731081326-phpapp01/85/Open-PHACTS-API-Walkthrough-32-320.jpg)