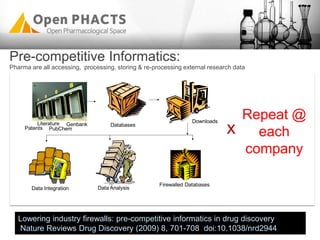
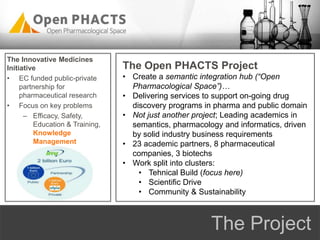
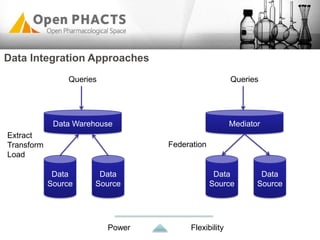
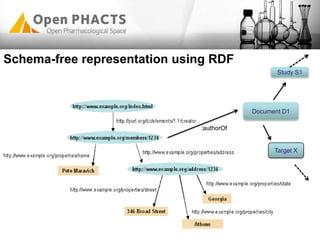
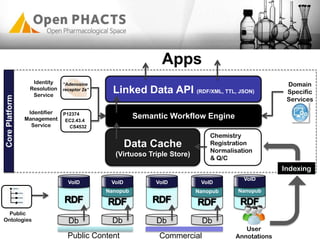
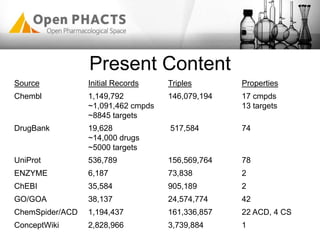
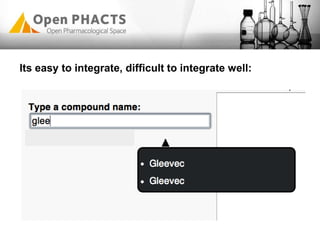
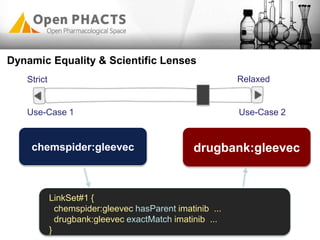
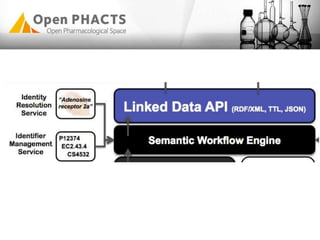
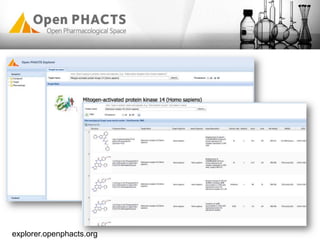
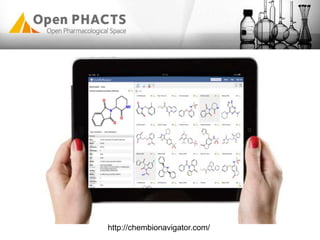
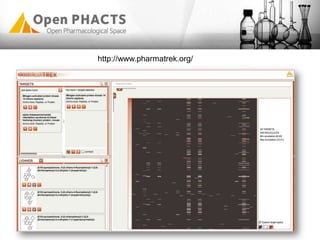
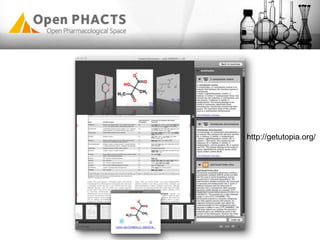
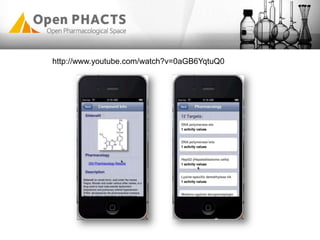
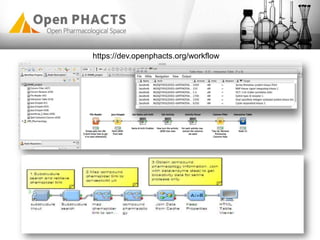
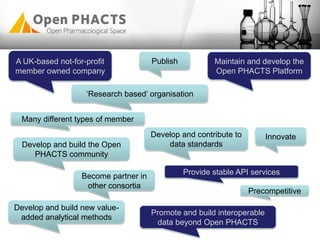
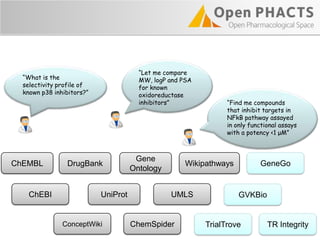
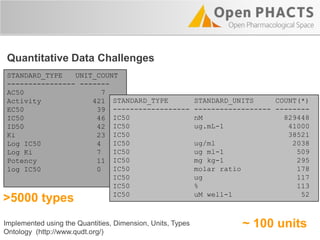
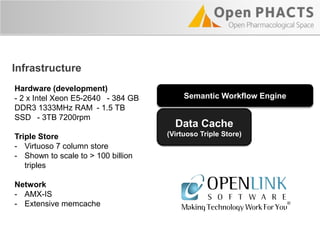
The Open PhaCTS project, a public-private partnership funded by the EC, aims to create a semantic integration hub for pharmaceutical research, facilitating data integration and analysis from various sources in drug discovery. With participation from 23 academic partners and 11 pharmaceutical companies, the project focuses on developing standards and tools to enhance data accessibility and improve pre-competitive informatics in the industry. The initiative promotes collaboration among stakeholders to foster innovation in drug development through an open API and community engagement.