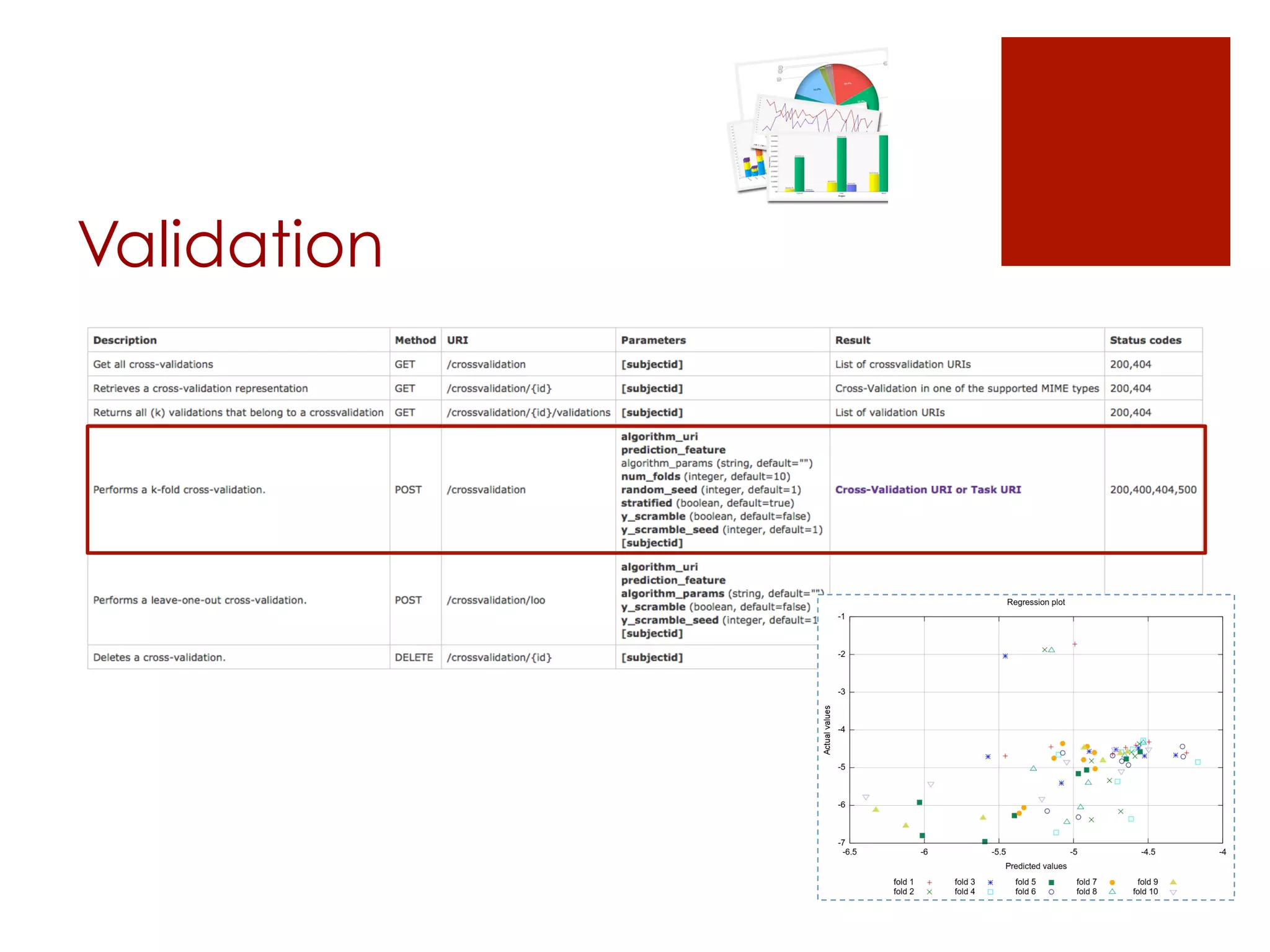
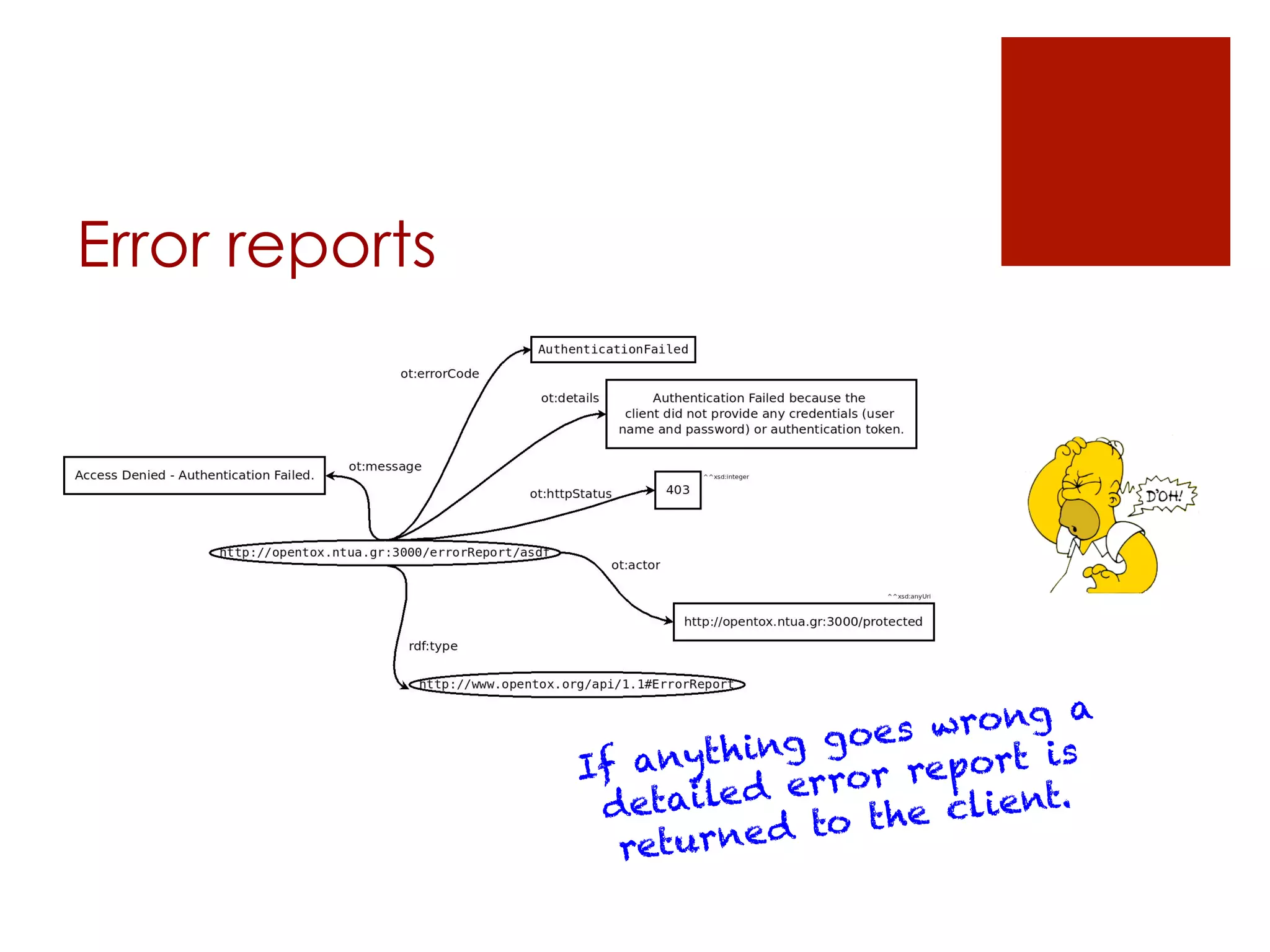
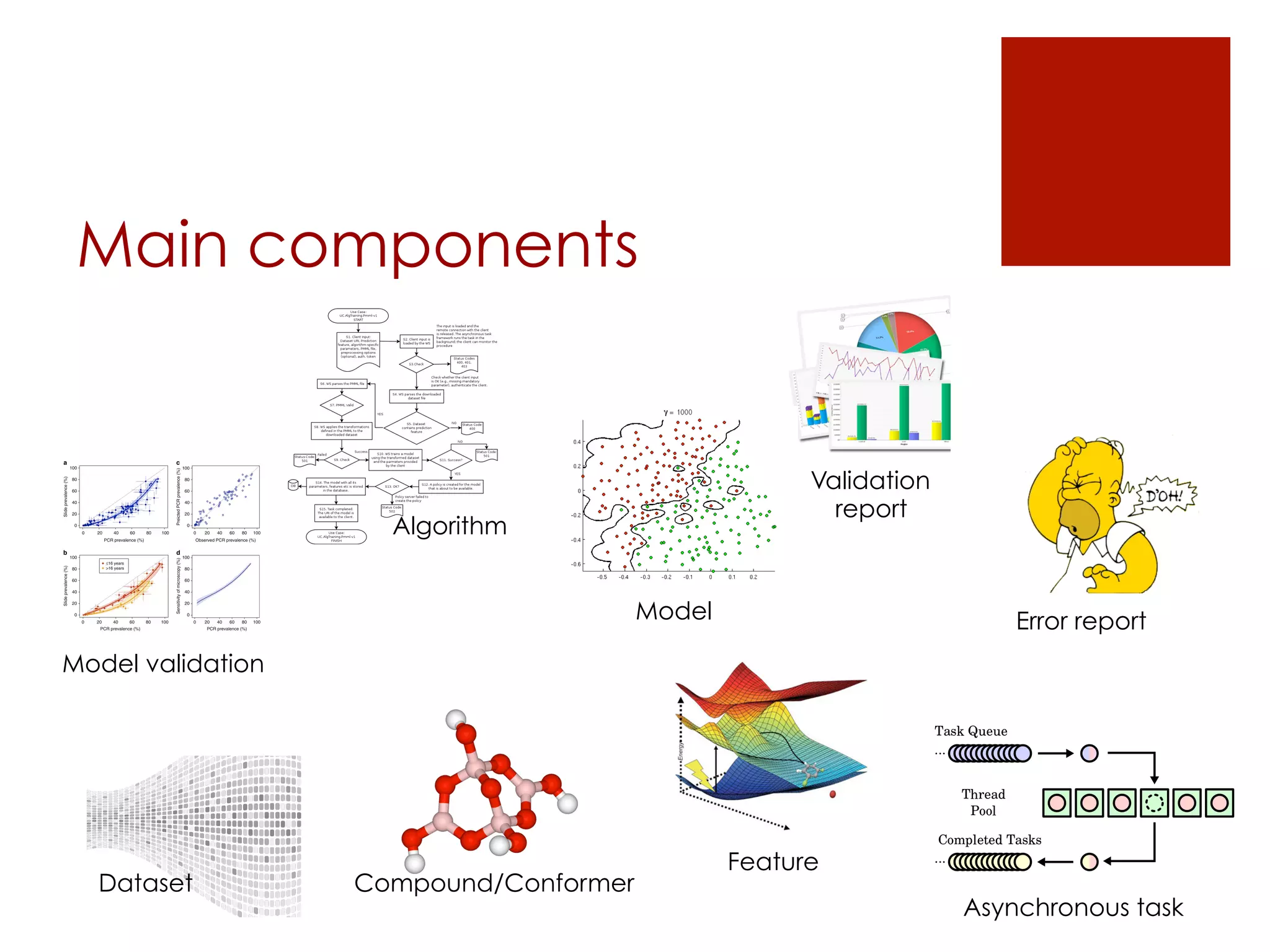
The document describes the OpenTox REST API, which provides a standardized way to access predictive toxicology web services and share data. It discusses the API's design principles, main components, access control policies, and features for working with data like compounds, conformers, datasets, algorithms, models, and validation reports. Examples using cURL requests are also provided to illustrate how the API can be used to perform tasks like creating a prediction job, monitoring its status, and retrieving results.
![OpenTox REST API
[A brief overview]
P. Sopasakis
1 National Technical University of Athens, Greece,
2 IMT Institute for Advanced Studies Lucca, Italy.](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-1-2048.jpg)
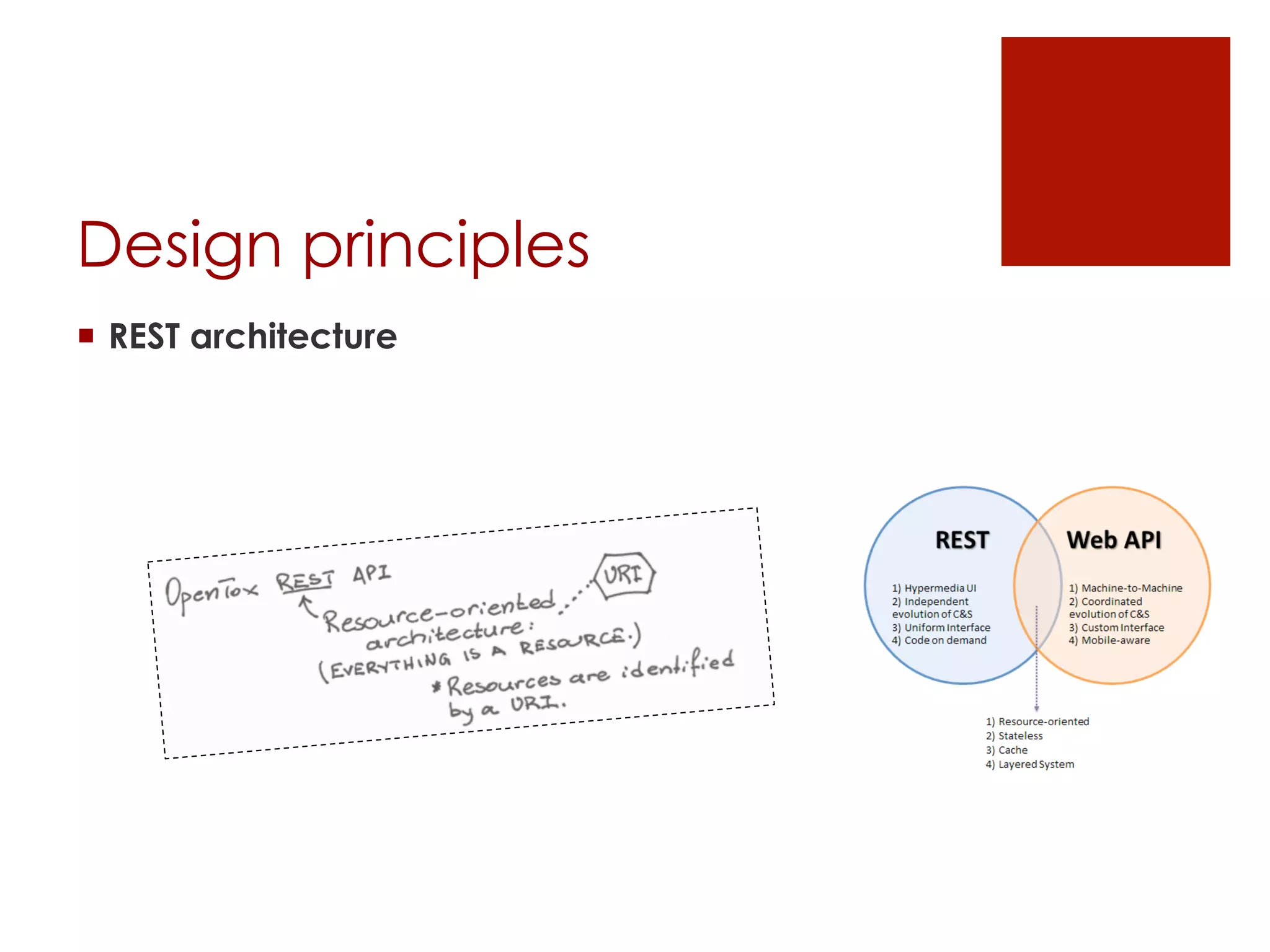
![Part I
The OpenTox framework
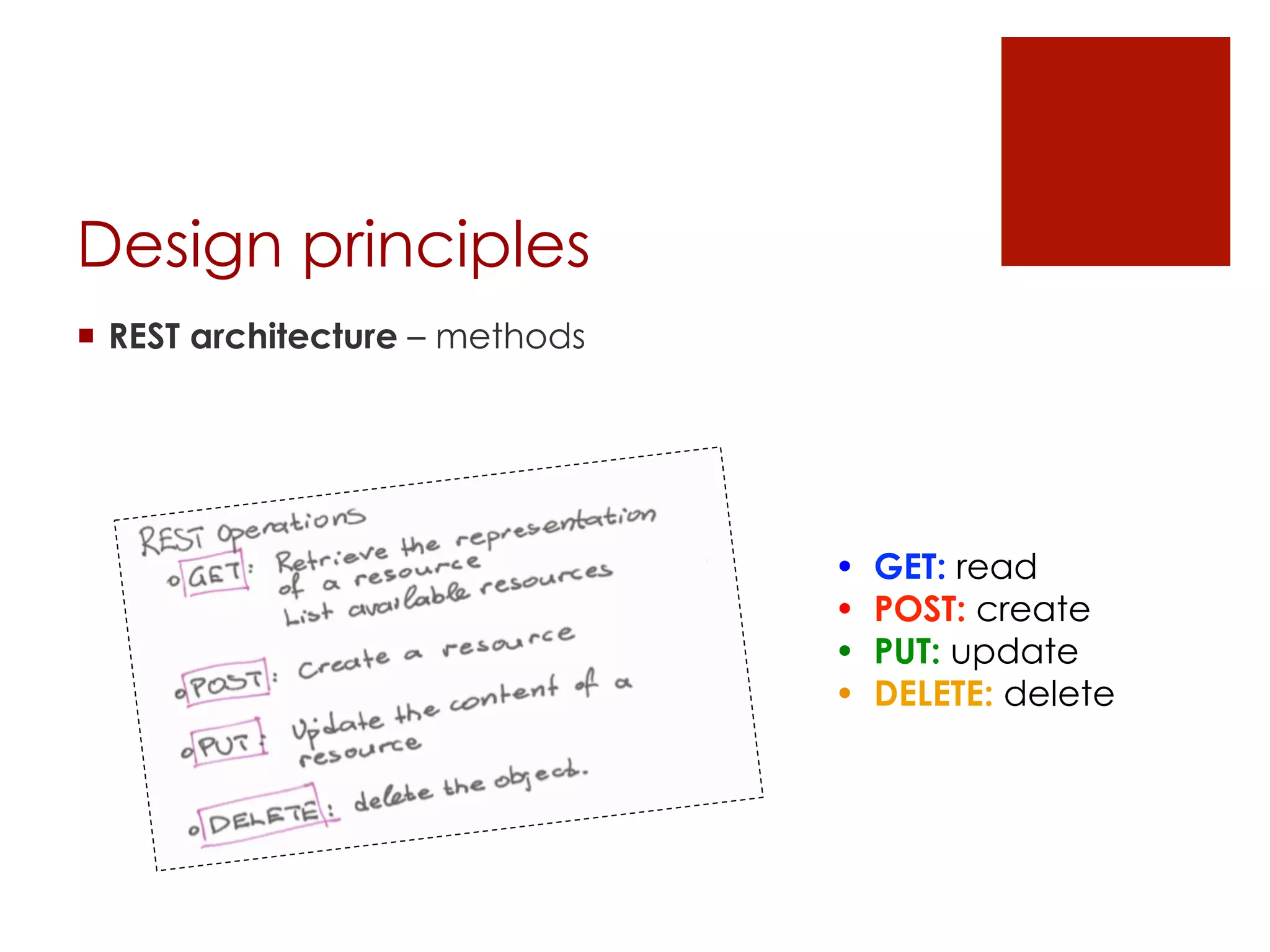
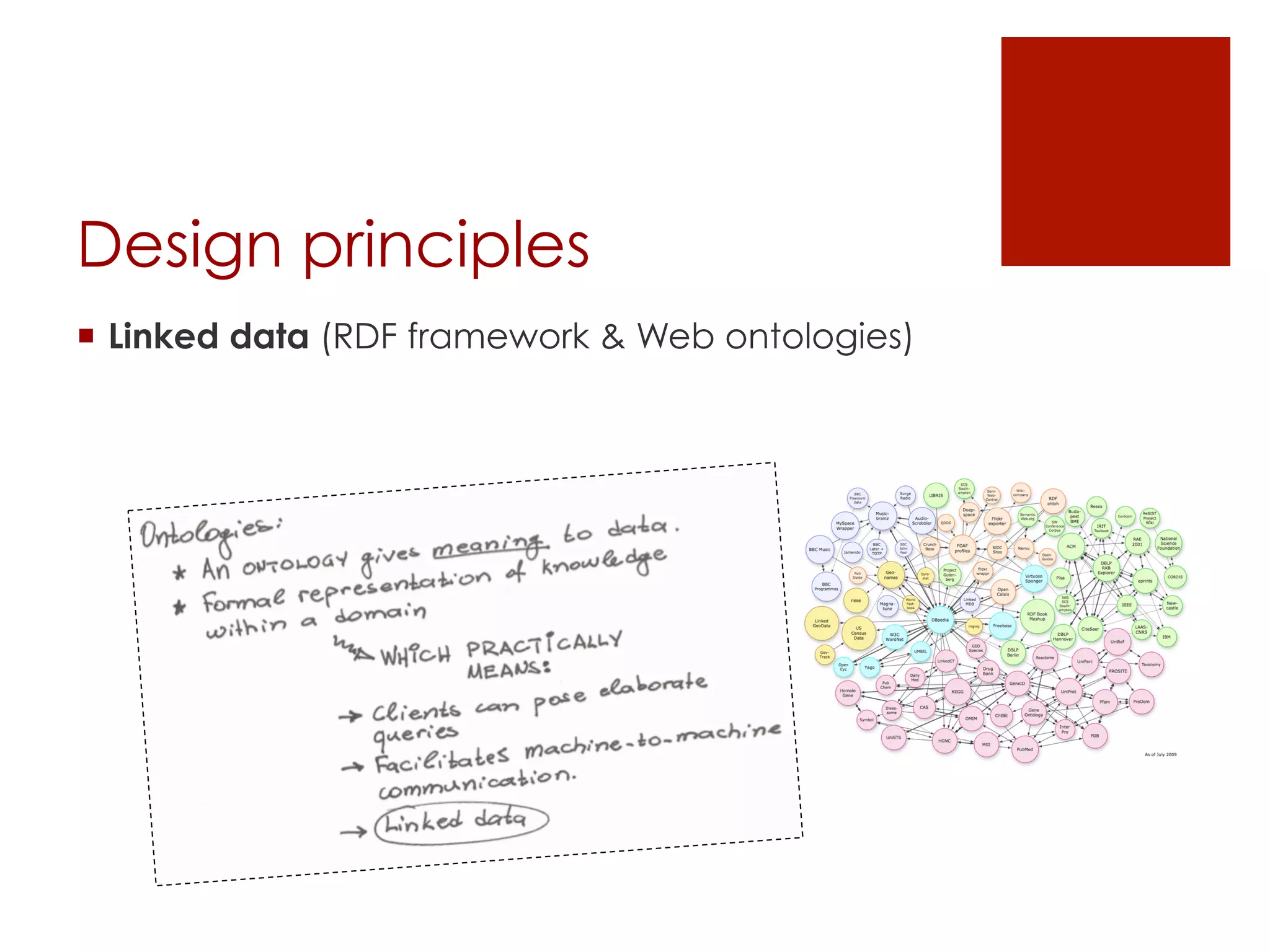
[design principles]
[components]
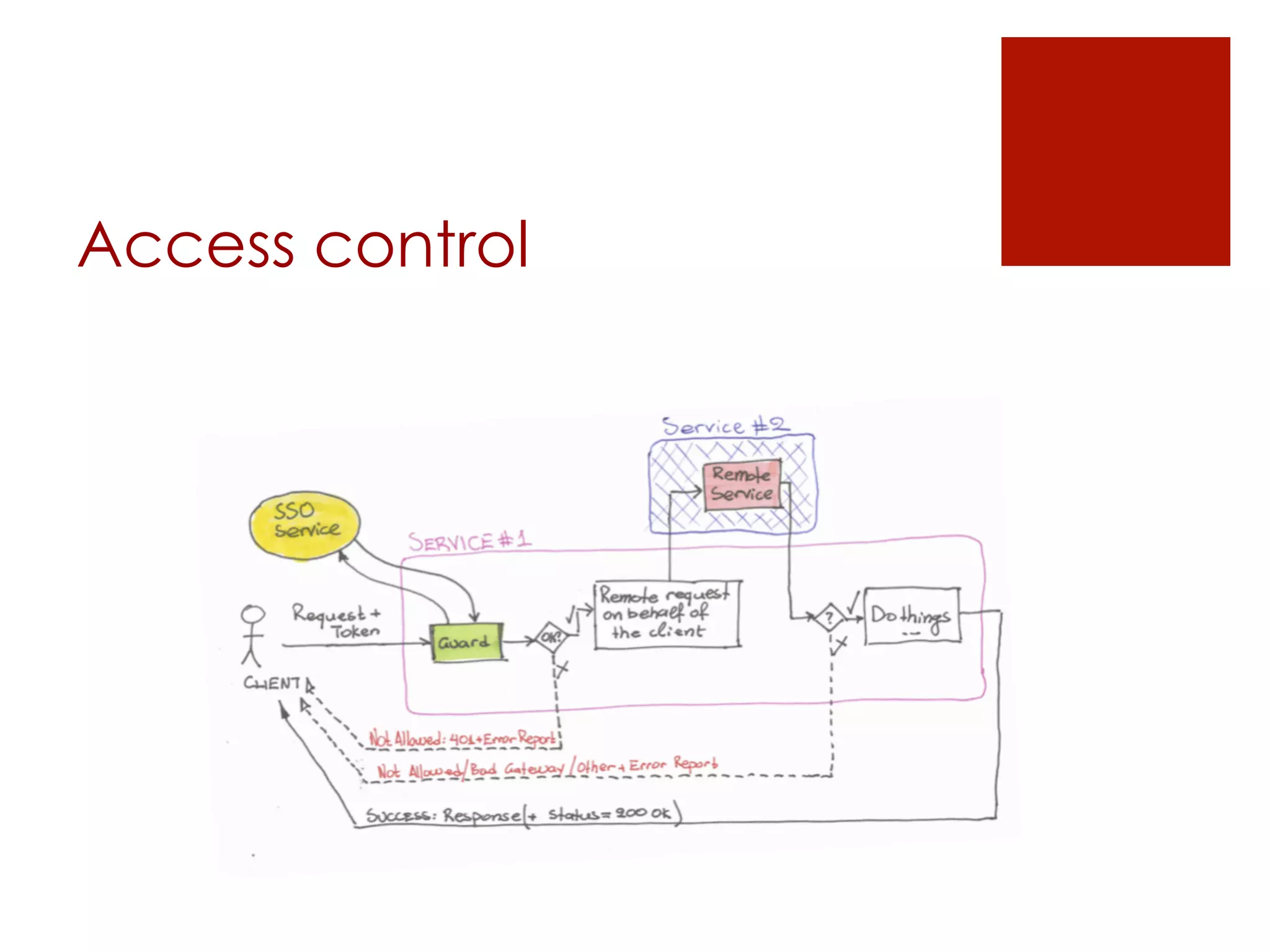
[access control]](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-2-2048.jpg)
![Part II
The OpenTox API
[with cURL examples]](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-9-2048.jpg)
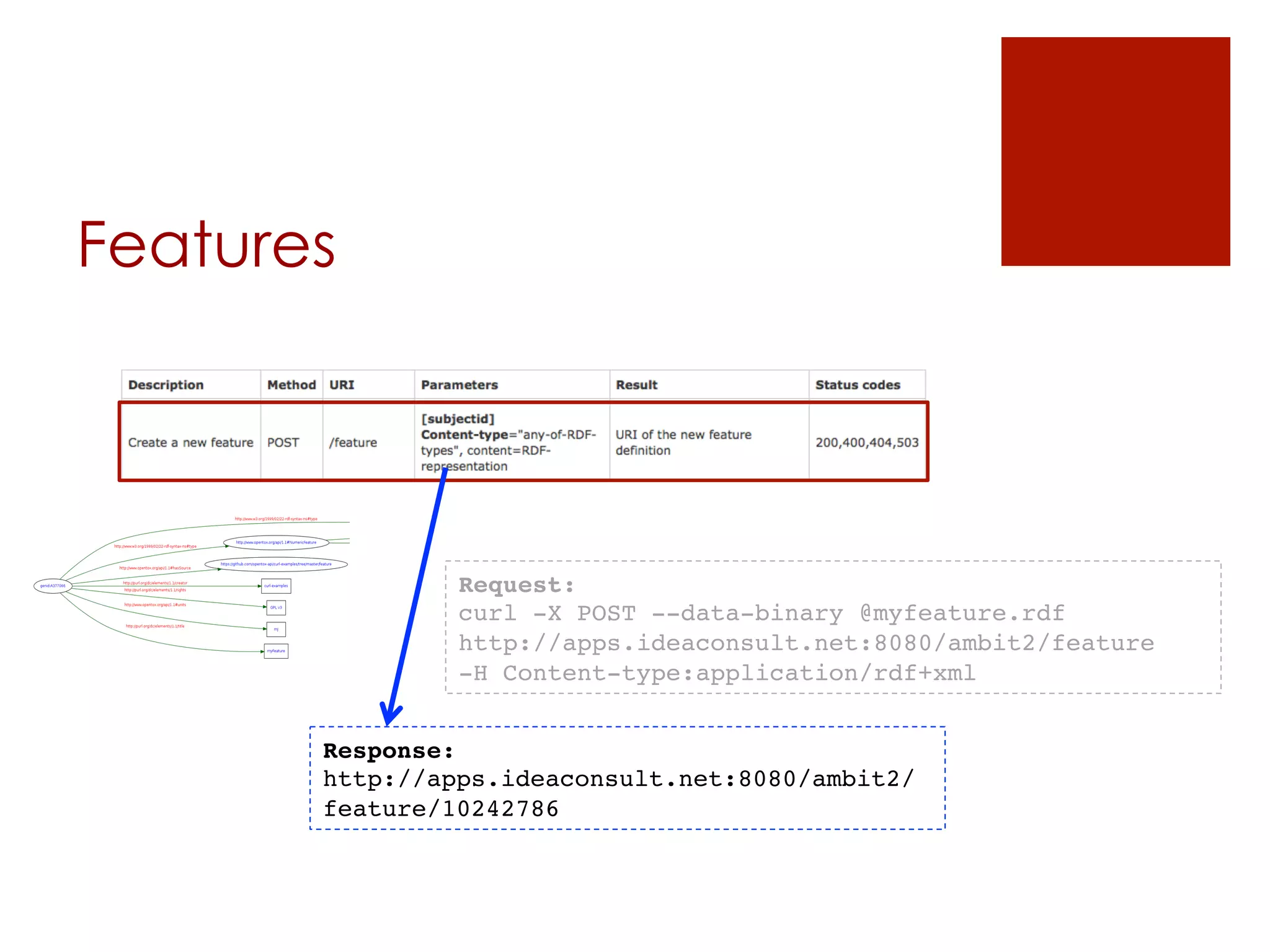
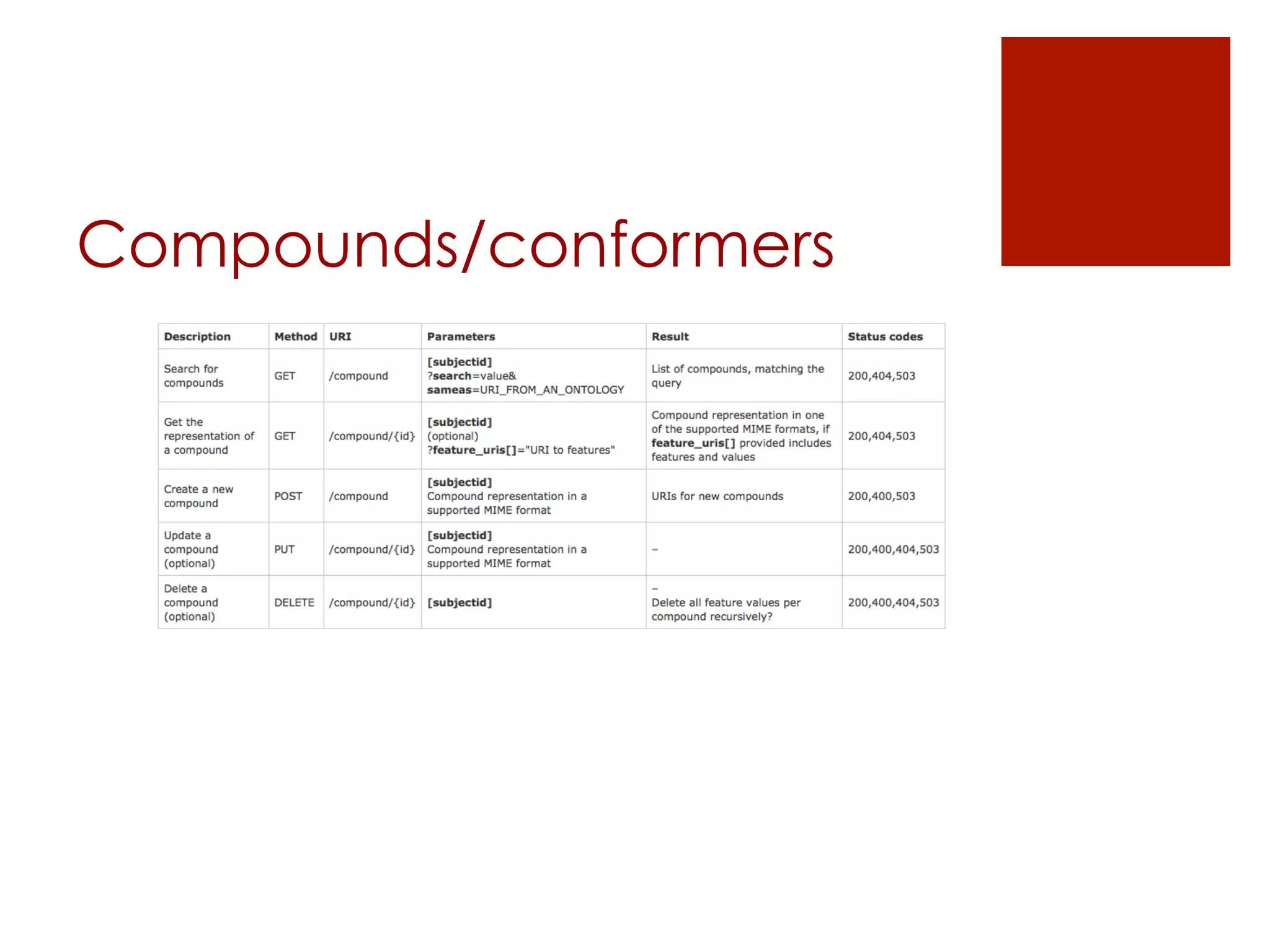
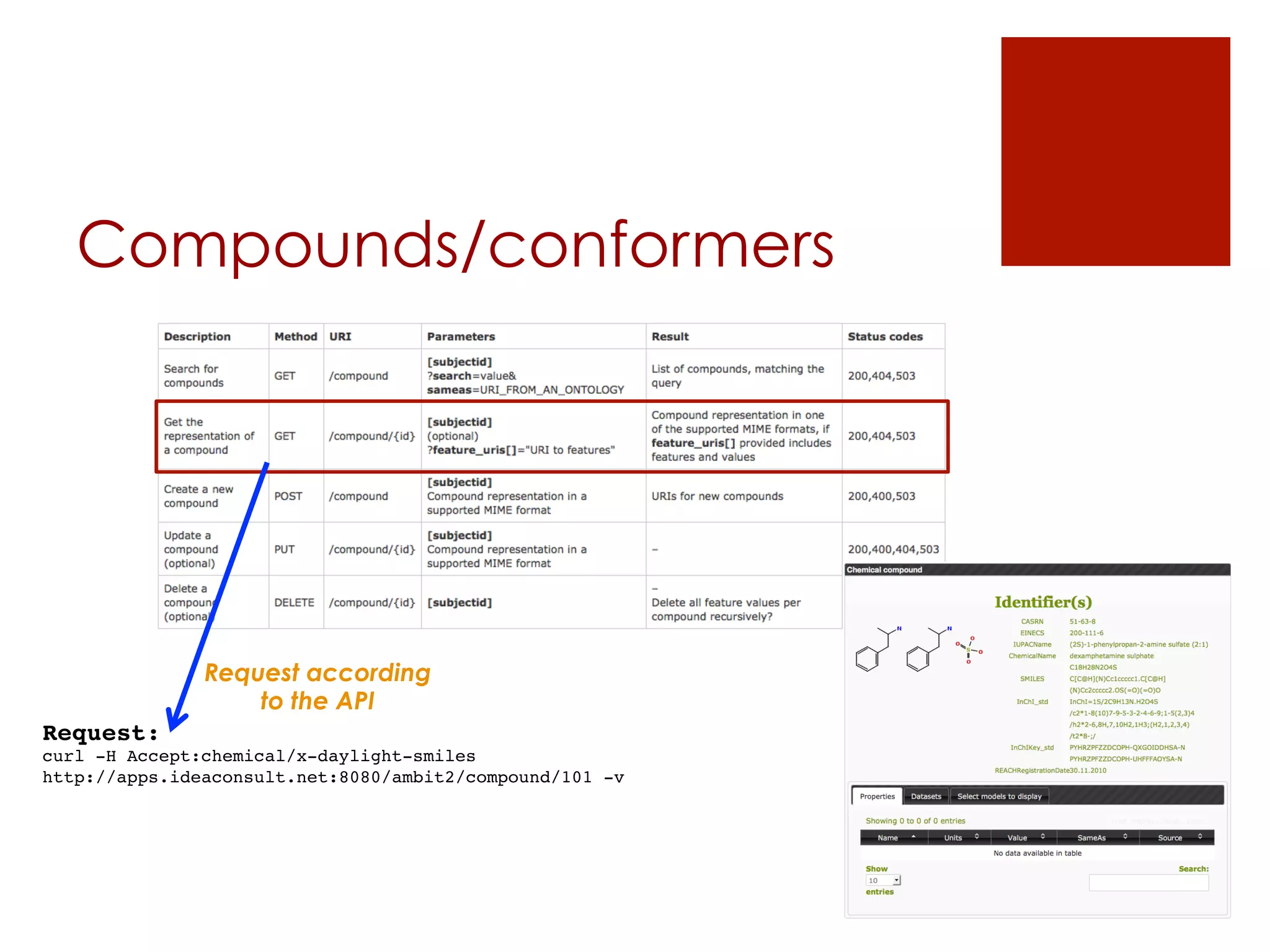
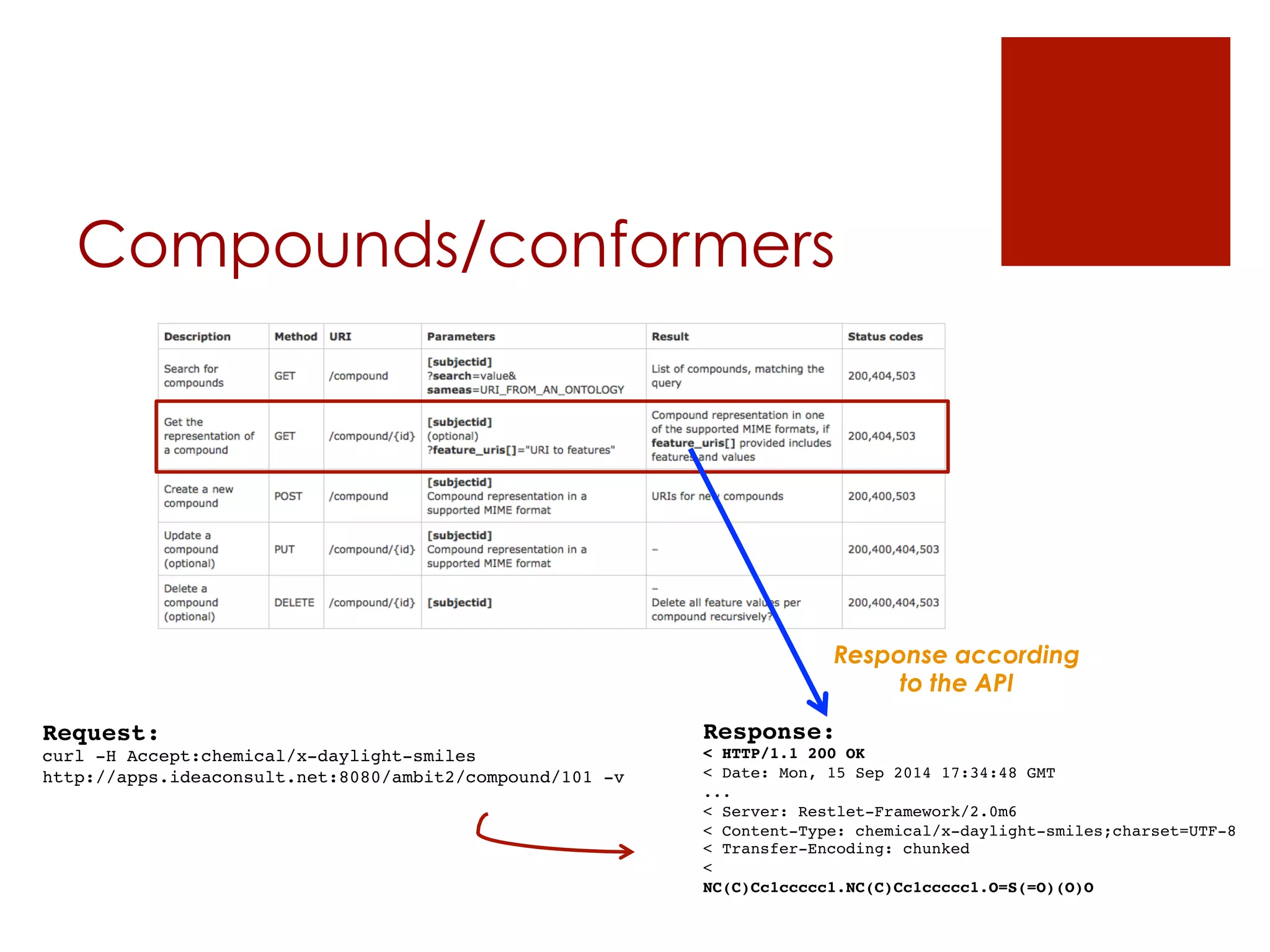

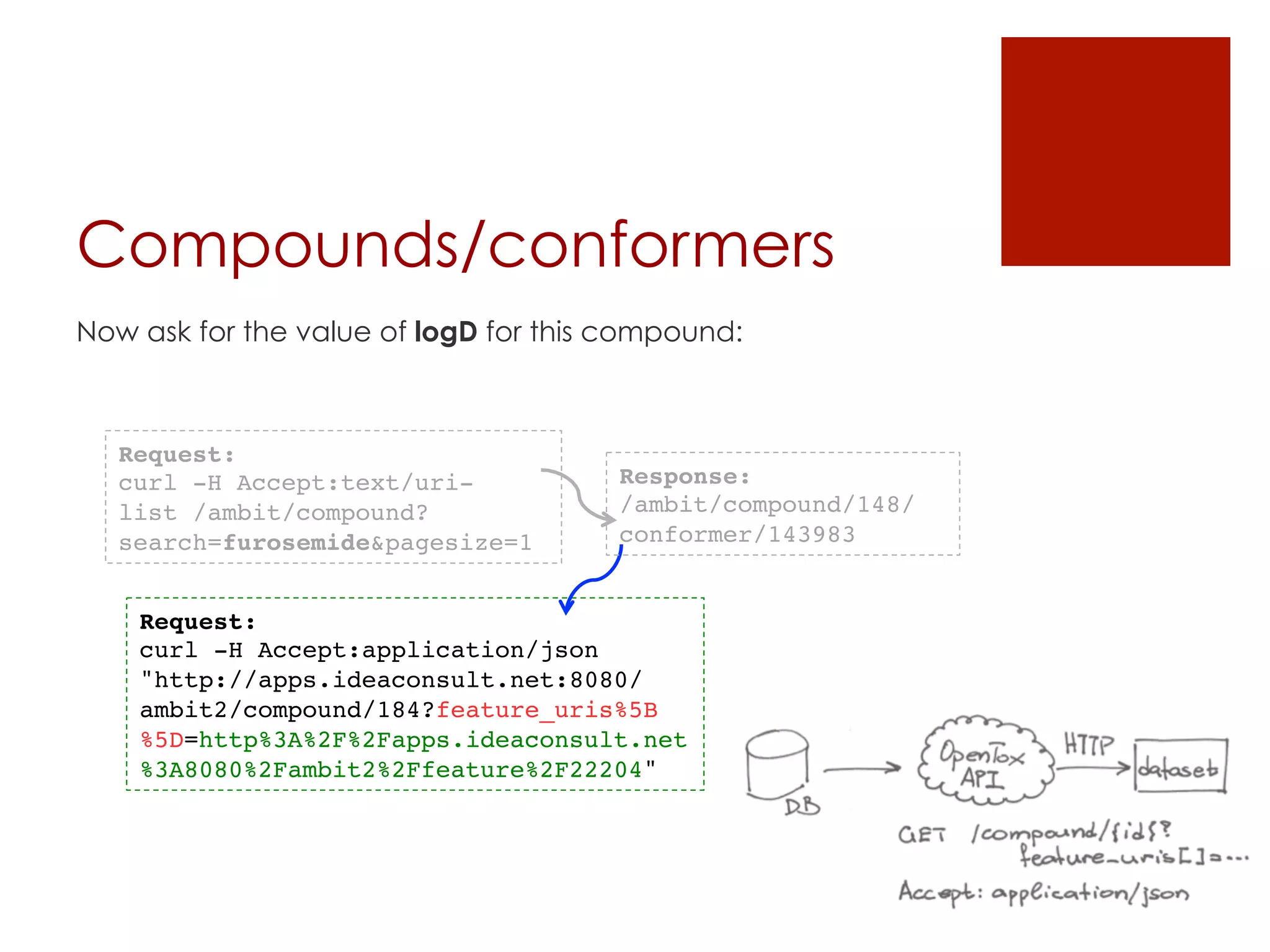
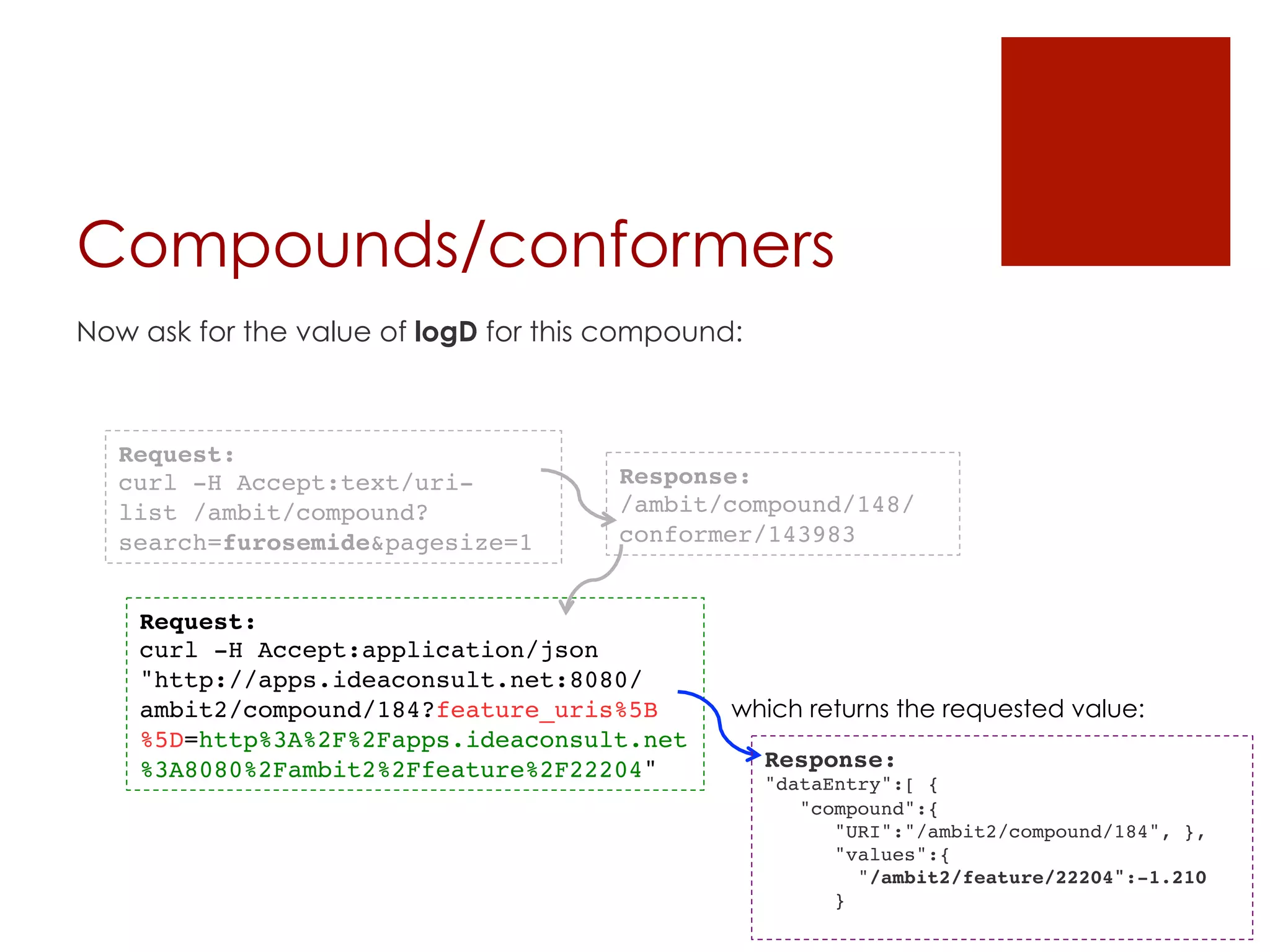
![Datasets
[API specs]
¡ GET:
¡ Download in RDF, JSON, CSV, ARFF or other formats, or get a
single feature-value pair for a compound,
¡ Query a dataset, select compounds or features
¡ Get metadata of the dataset
¡ POST:
¡ Create a new dataset on the server,
¡ PUT:
¡ Update the dataset
¡ DELETE:
¡ Delete a dataset,
¡ Delete part of a dataset (some compounds and/or features)](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-20-2048.jpg)
![Datasets
[RDF representation]
* this structure is determined and understood by
the OpenTox ontology.](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-21-2048.jpg)
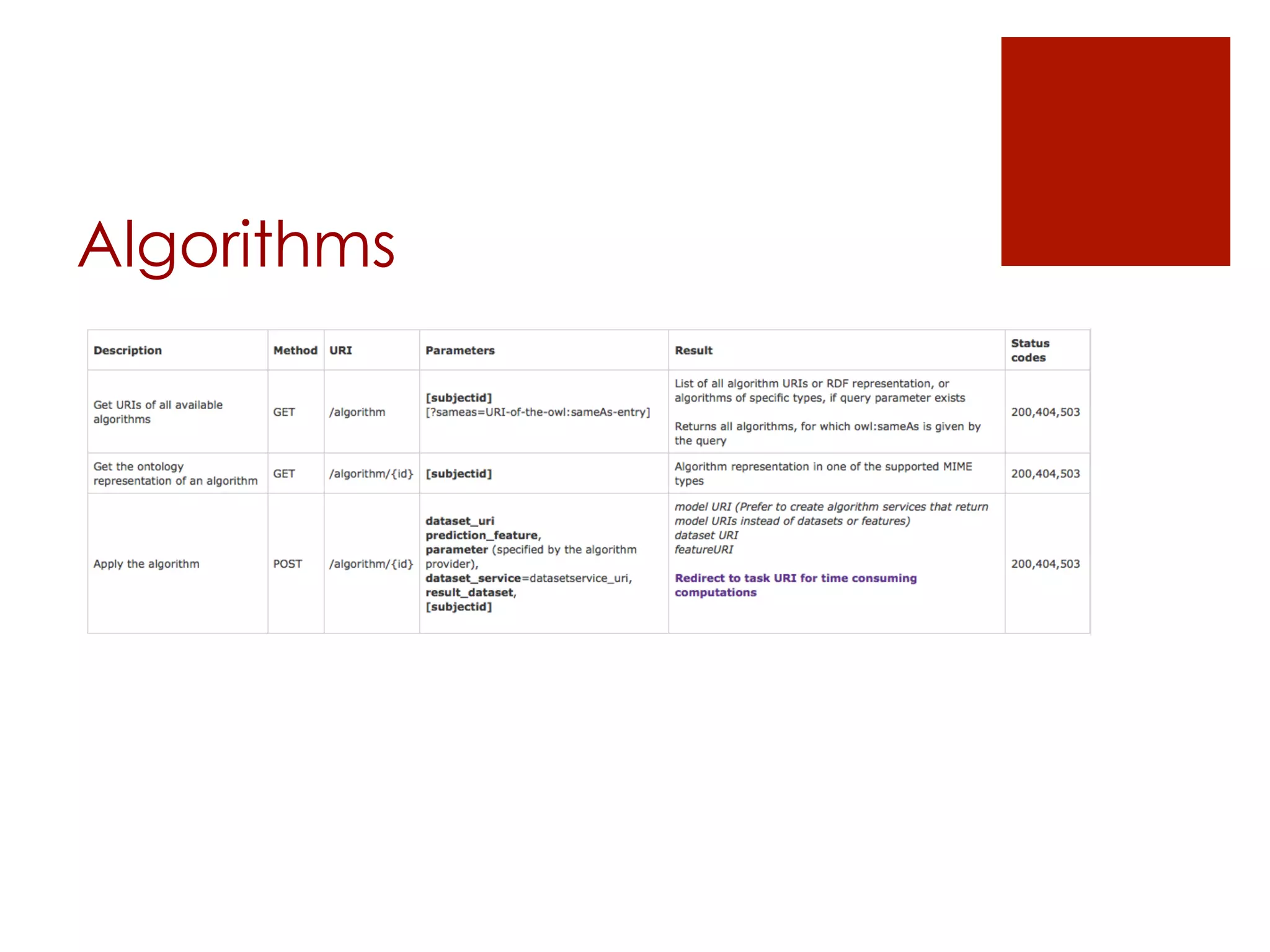
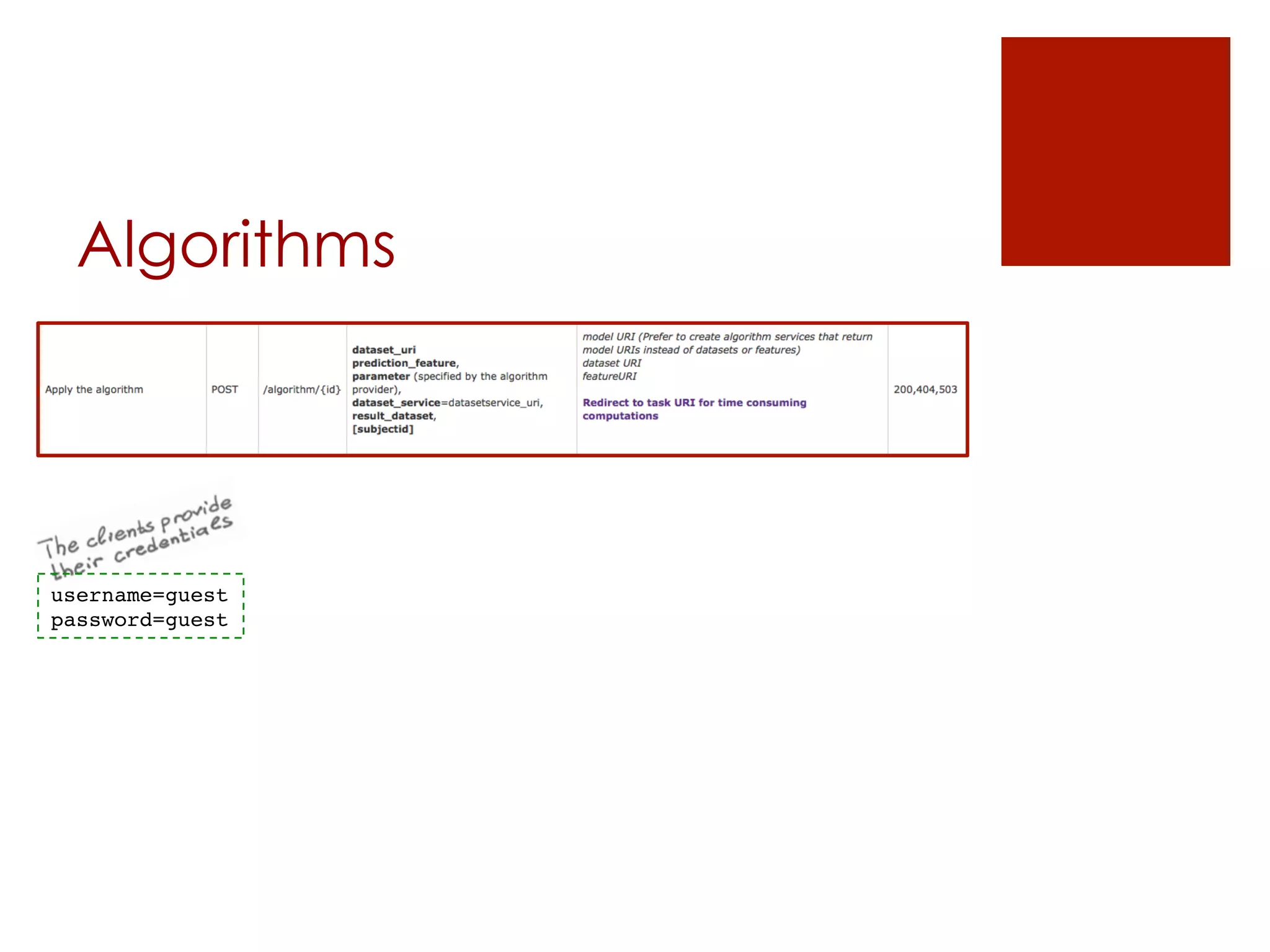



![Algorithms
[Task API]](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-27-2048.jpg)
![Algorithms
[Task API]
while [ $status -eq 202 ]!
do!
sleep 2!
status=$(curl --write-out %{http_code} !
--silent --output /dev/null $task_uri)!
done!](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-28-2048.jpg)
![Algorithms
[Task API]
while [ $status -eq 202 ]!
do!
sleep 2!
status=$(curl --write-out %{http_code} !
--silent --output /dev/null $task_uri)!
done!
model_uri=`curl -H subjectid:
$token -H Accept:text/uri-list
$task_uri 2> /dev/null`!](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-29-2048.jpg)
![Algorithms
[PMML Transformations]
Example request:!
curl -X POST -v -include !
--form pmml=@mypmml-transformation.xml !
--form epsilon=0.03 !
--form kernel=rbf !
-H Content-Type:multipart/form-data !
http://opentox.ntua.gr:8080/algorithm/svm!](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-30-2048.jpg)
![Models
[Export]
The PMML format is available for some
models (planning to provide full support
in the future) for the sake of portability
and transparency.](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-31-2048.jpg)
![Models
[Predict]
Models are used to produce predictions
given a dataset or a compound. A POST
is used to perform such predictions.](https://image.slidesharecdn.com/opentoxapiathens2014-140921205304-phpapp02/75/OpenTox-API-introductory-presentation-32-2048.jpg)