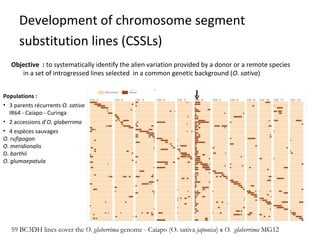
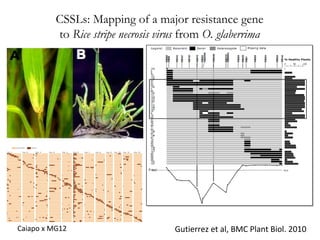
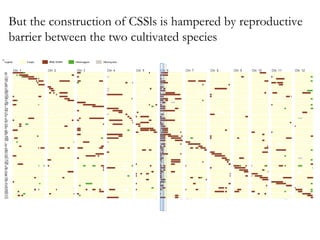
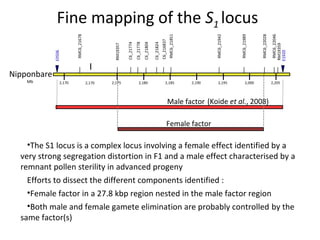
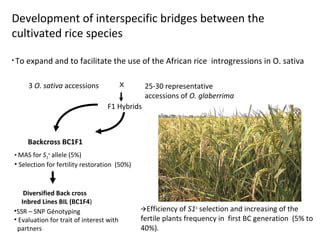
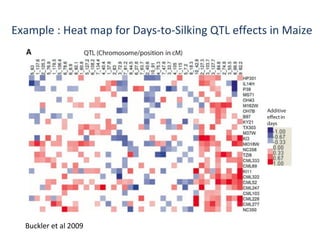
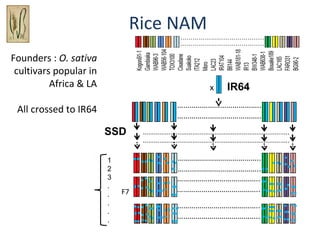
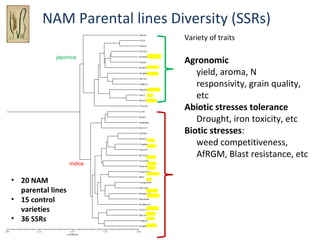
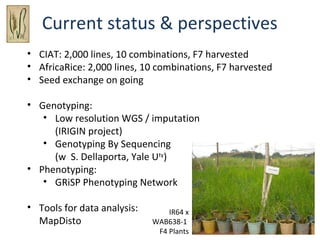
The document discusses innovative approaches for gene discovery and breeding in rice, focusing on the utilization of wild rice species to introduce valuable alleles into cultivated rice, notably through the development of chromosome segment substitution lines (CSSLs). It highlights the challenges posed by reproductive barriers between different rice species, specifically the s1 locus affecting fertility. Additionally, the document outlines the progress in developing interspecific bridges and nested association mapping to enhance drought tolerance and other traits in rice varieties.