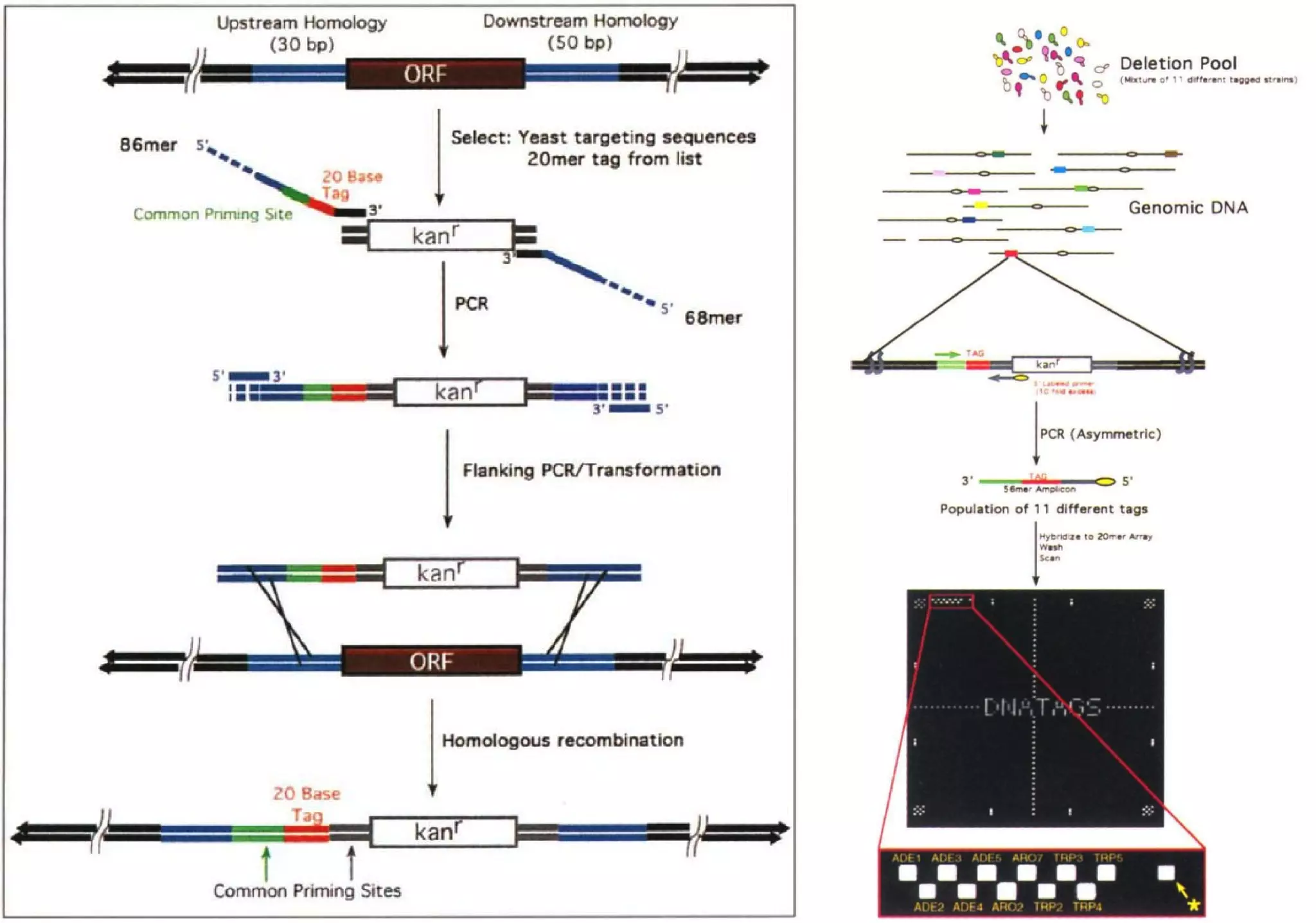
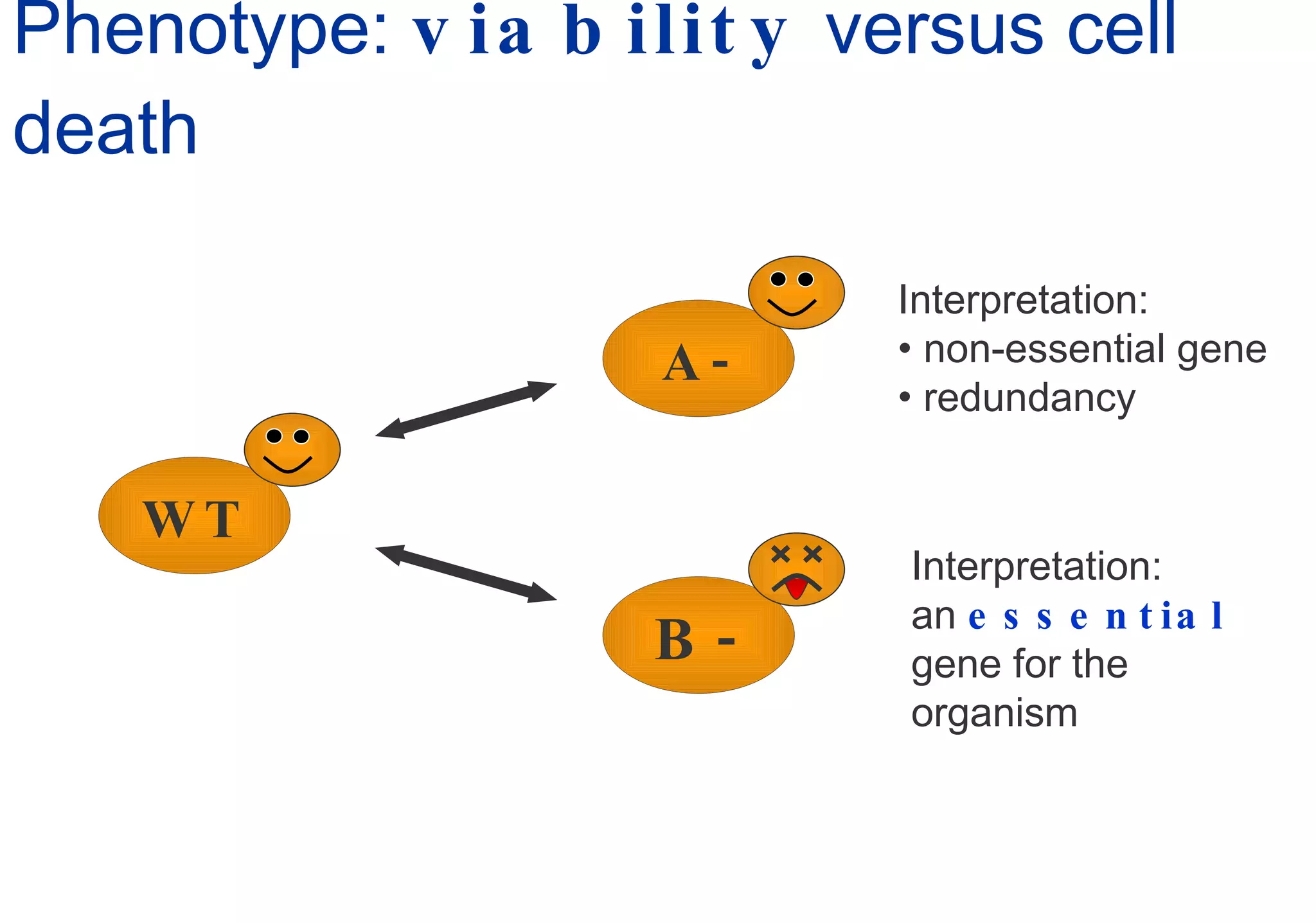
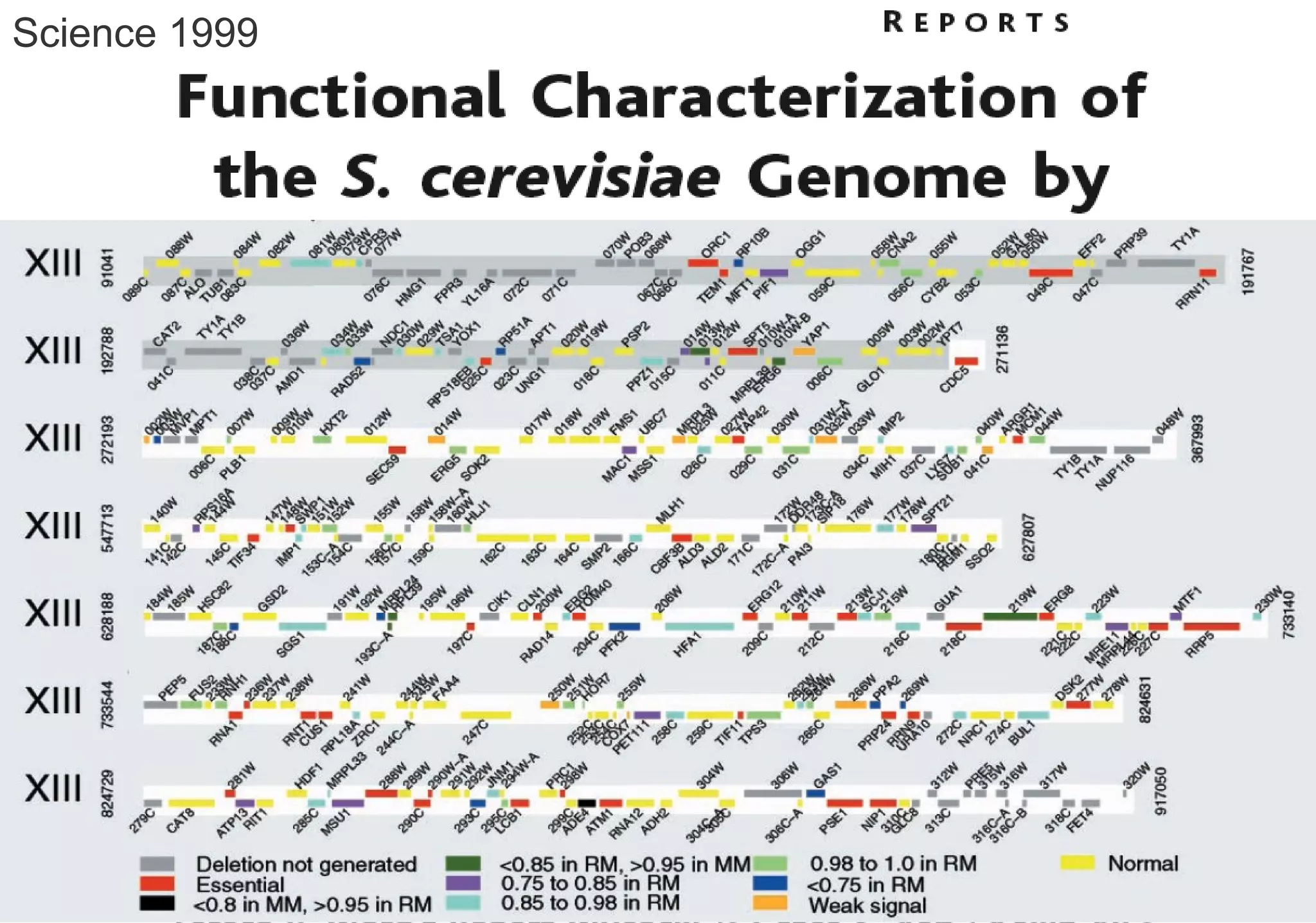
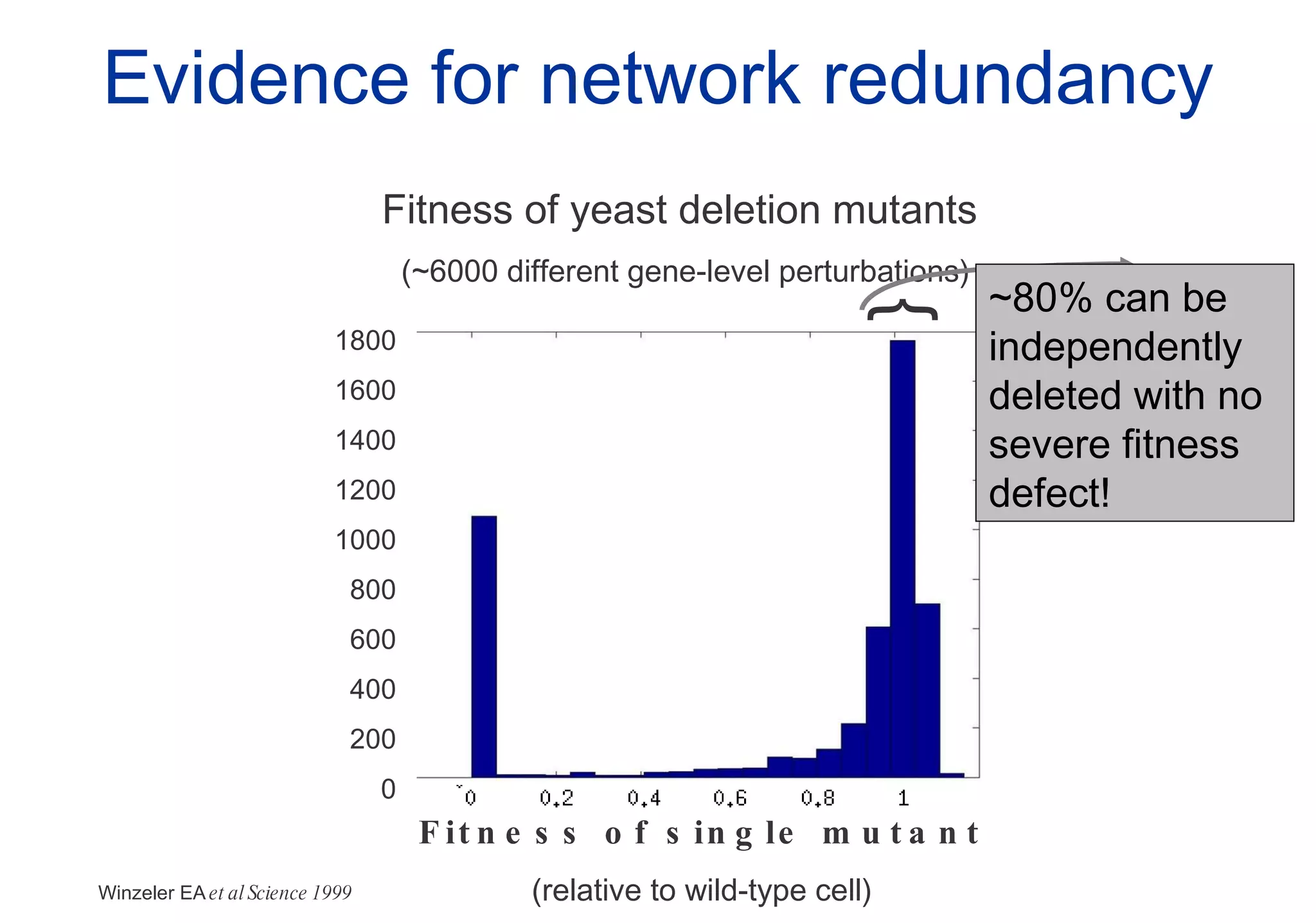
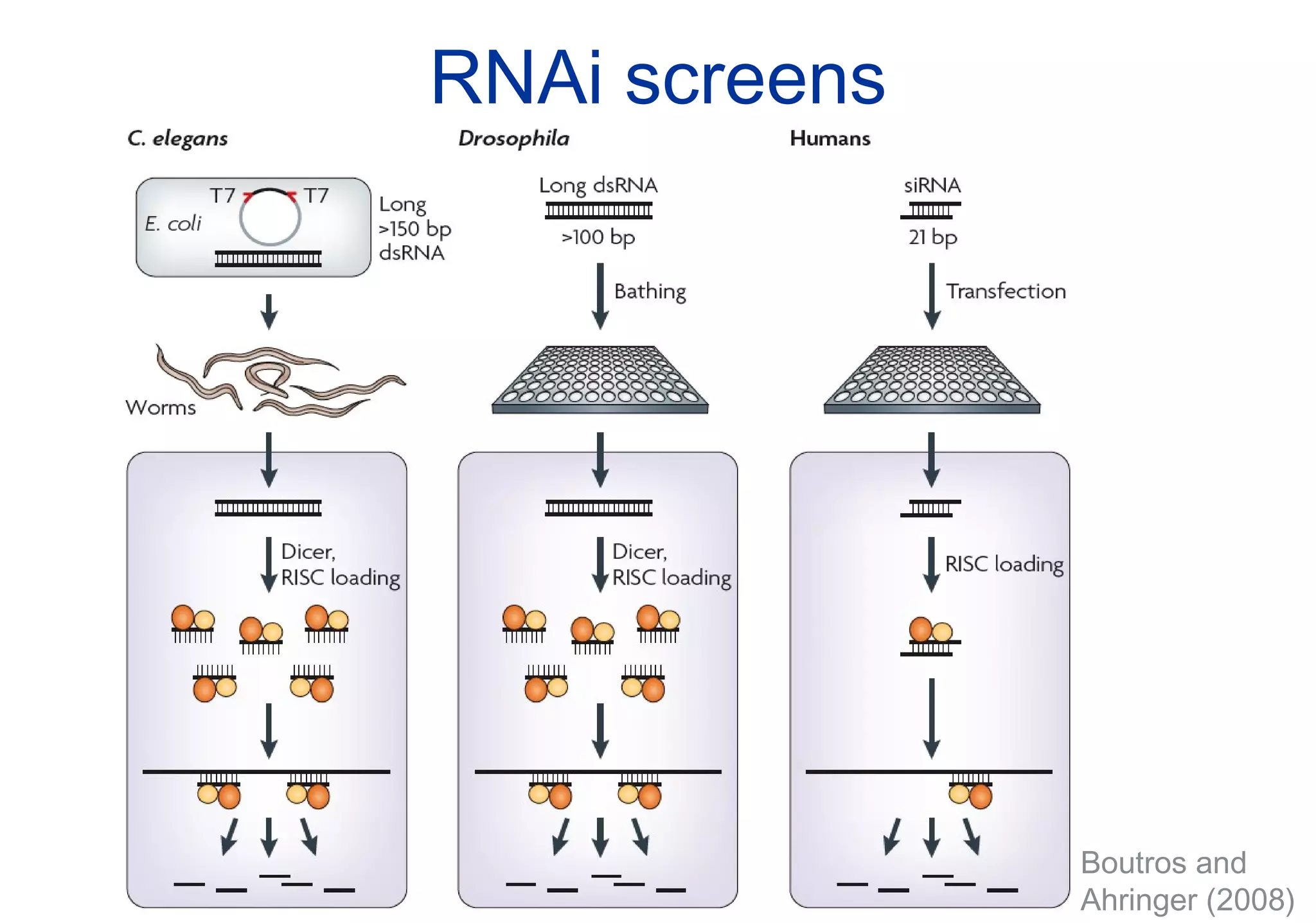
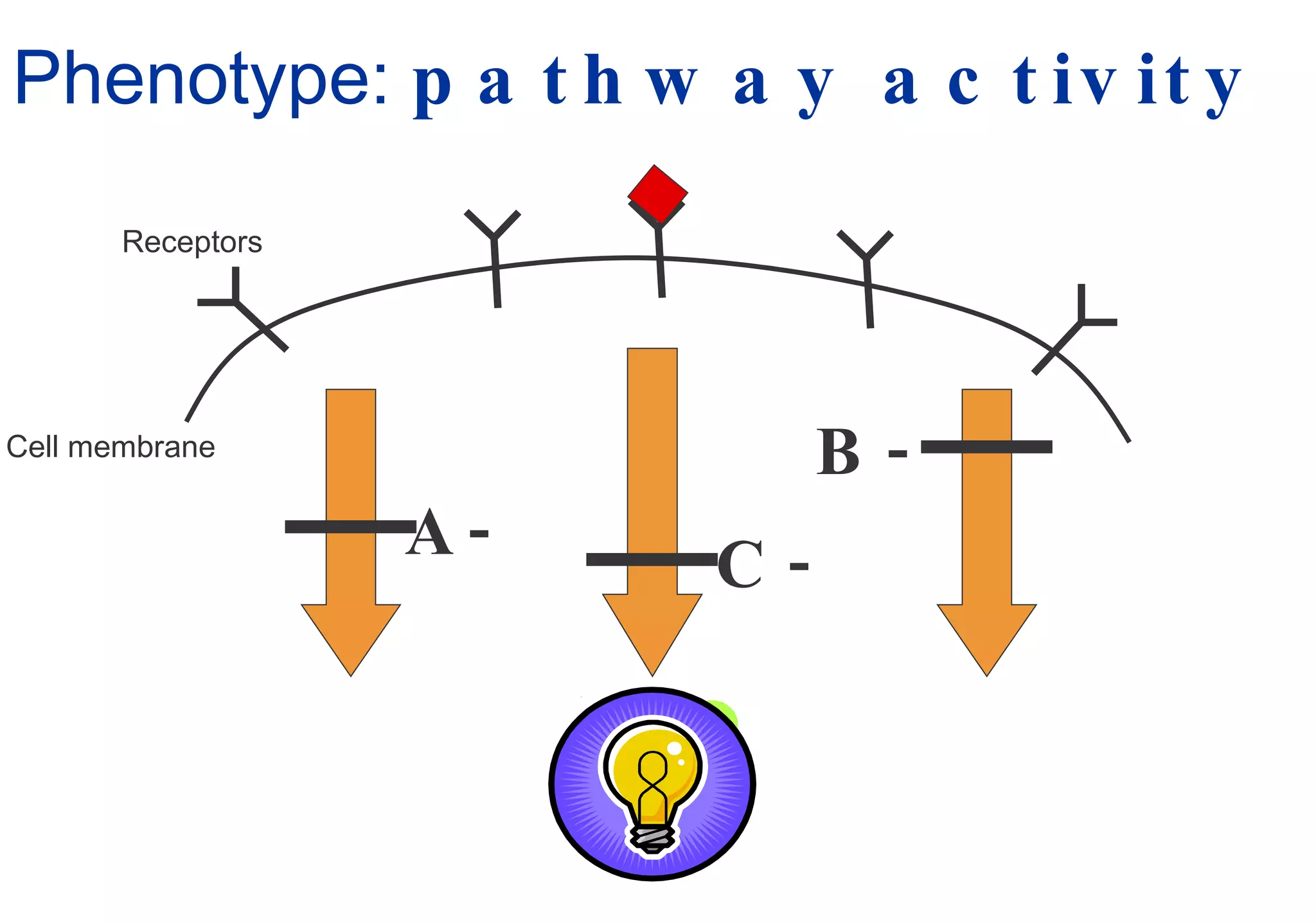
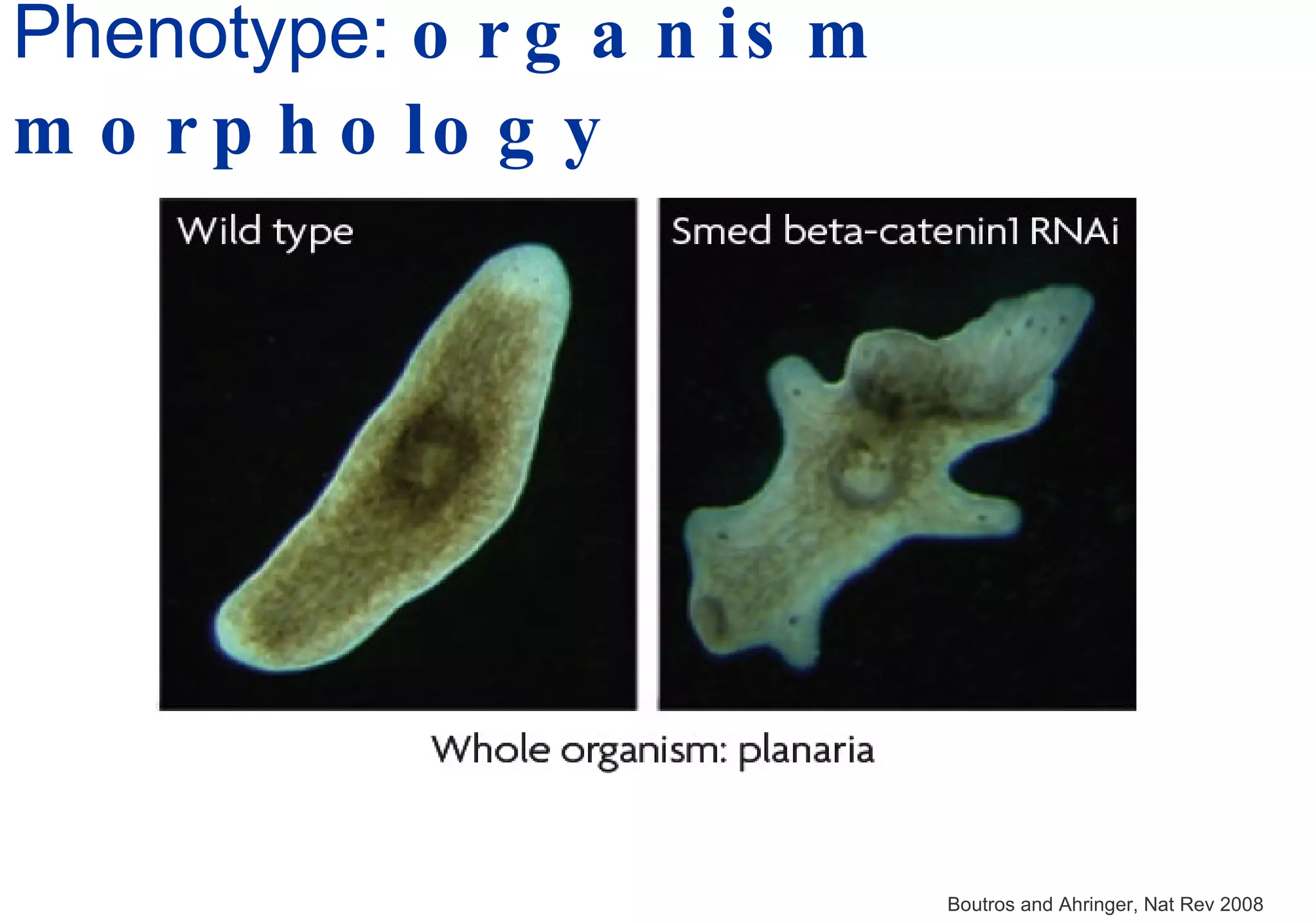
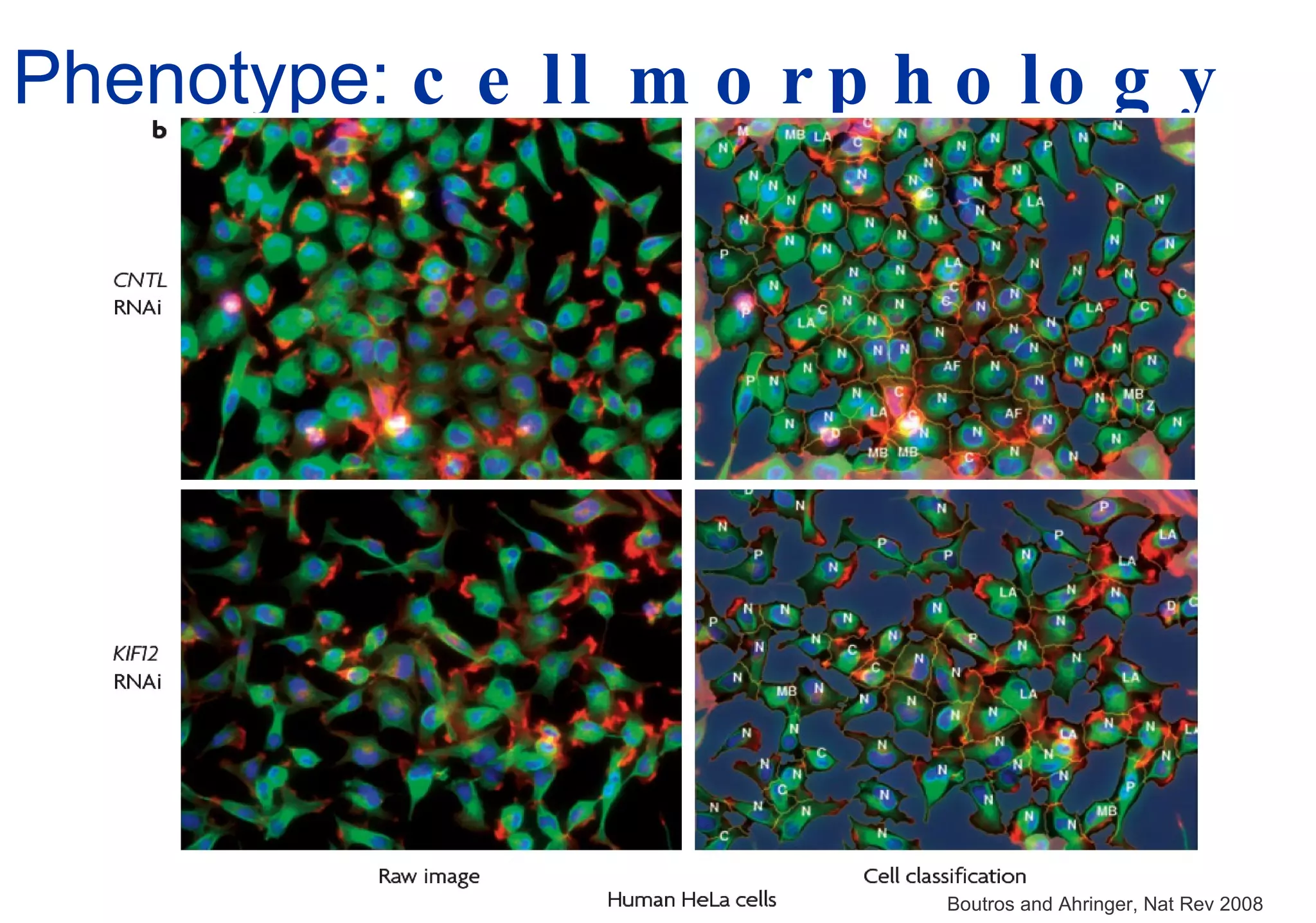
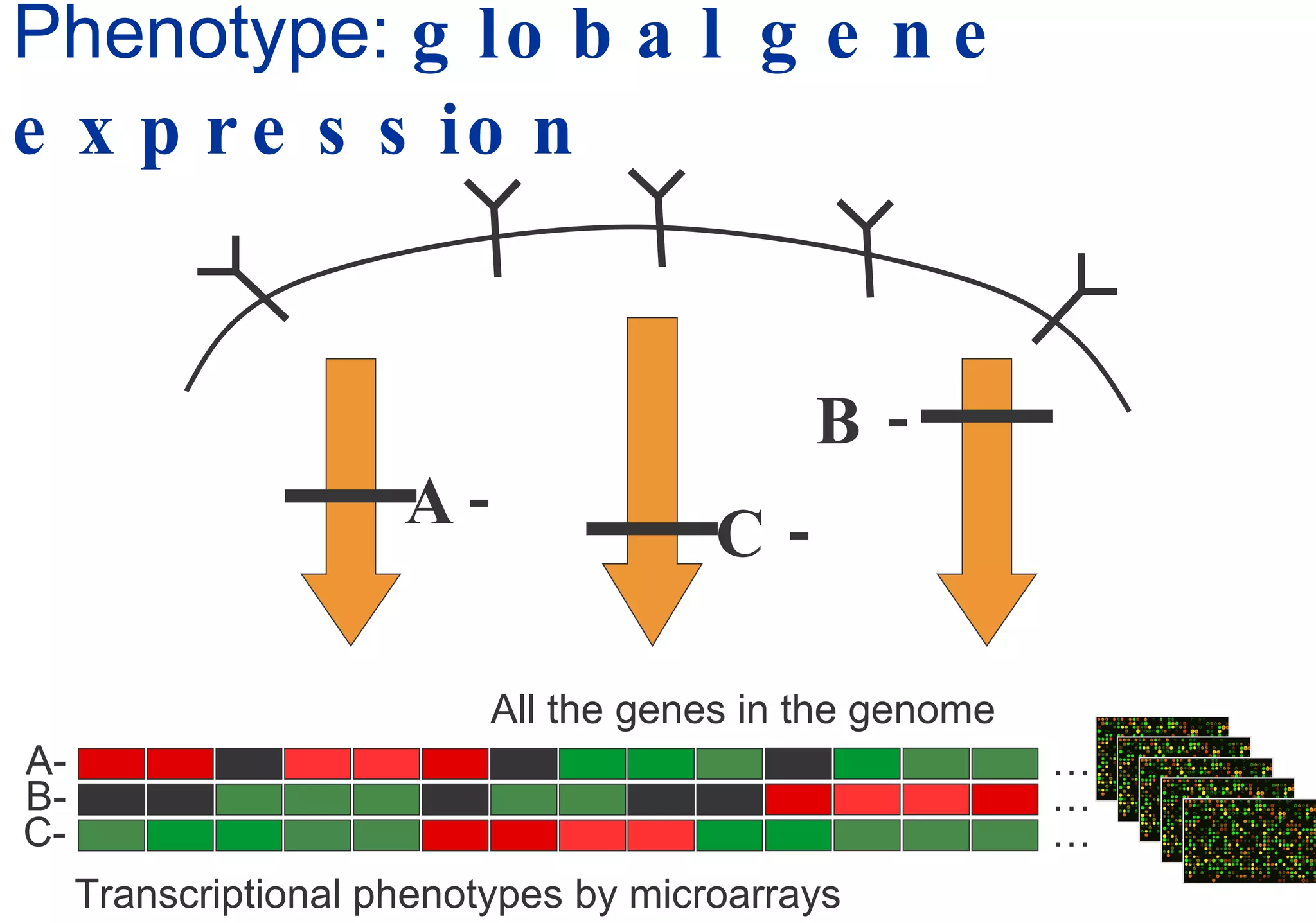
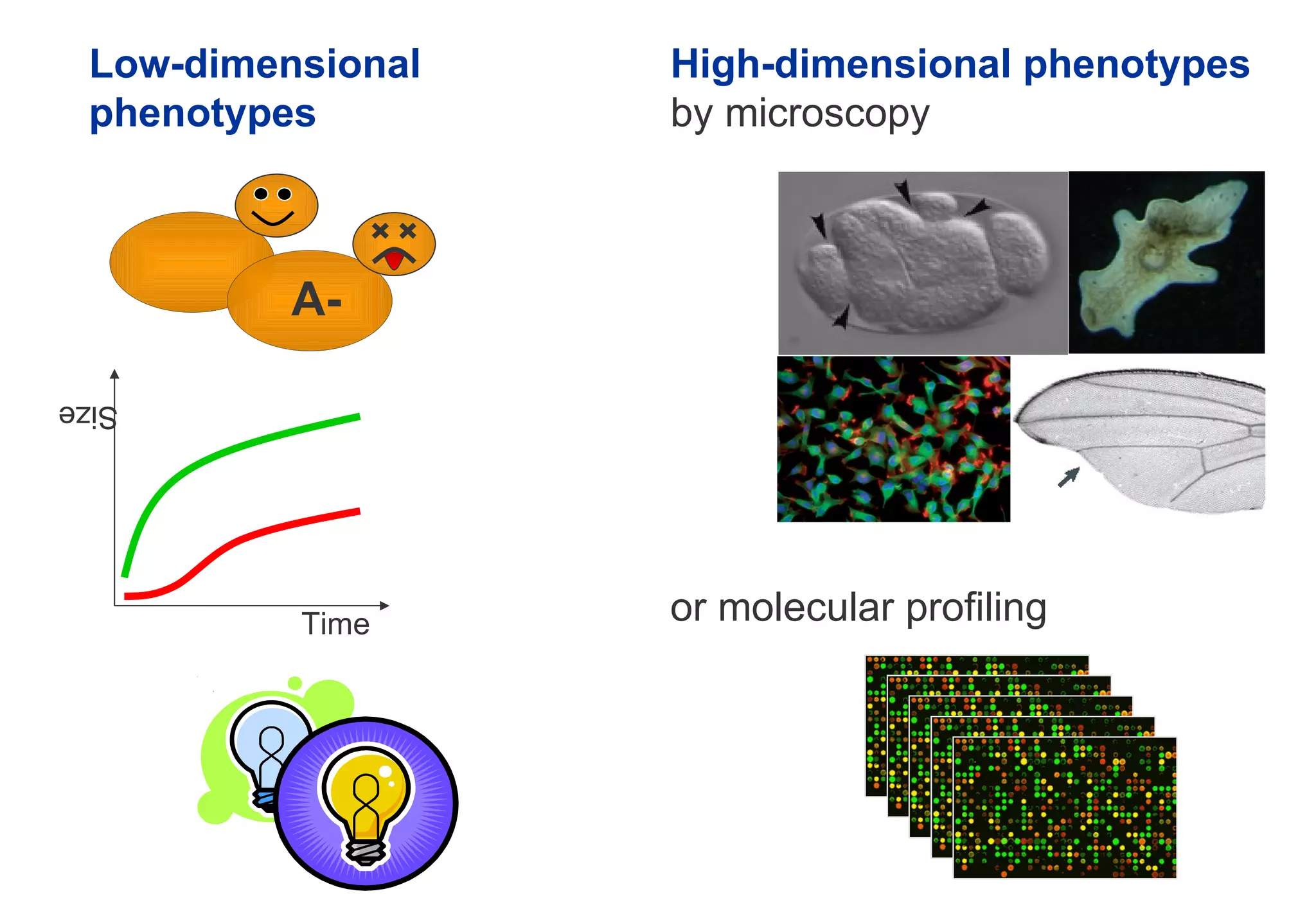
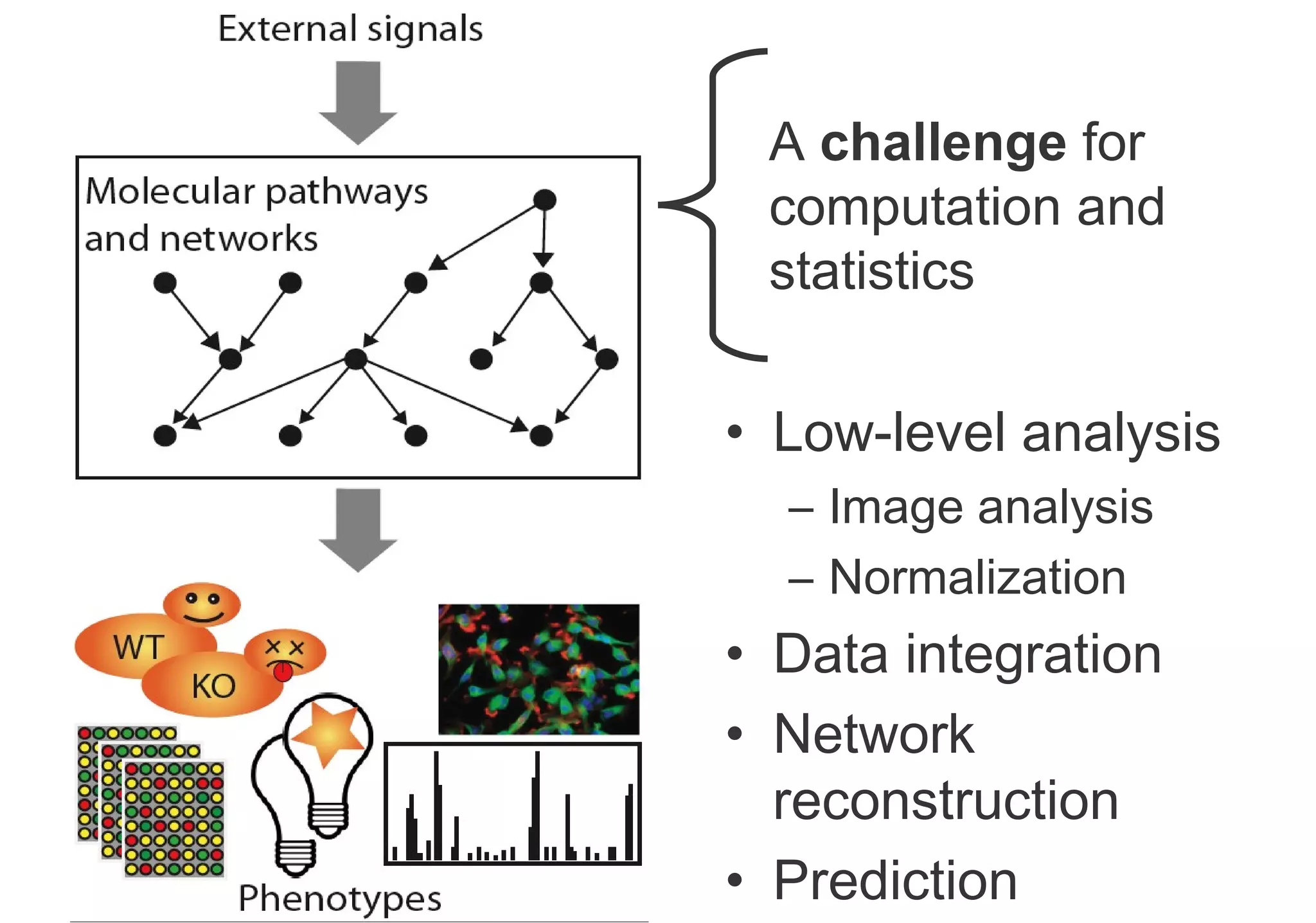
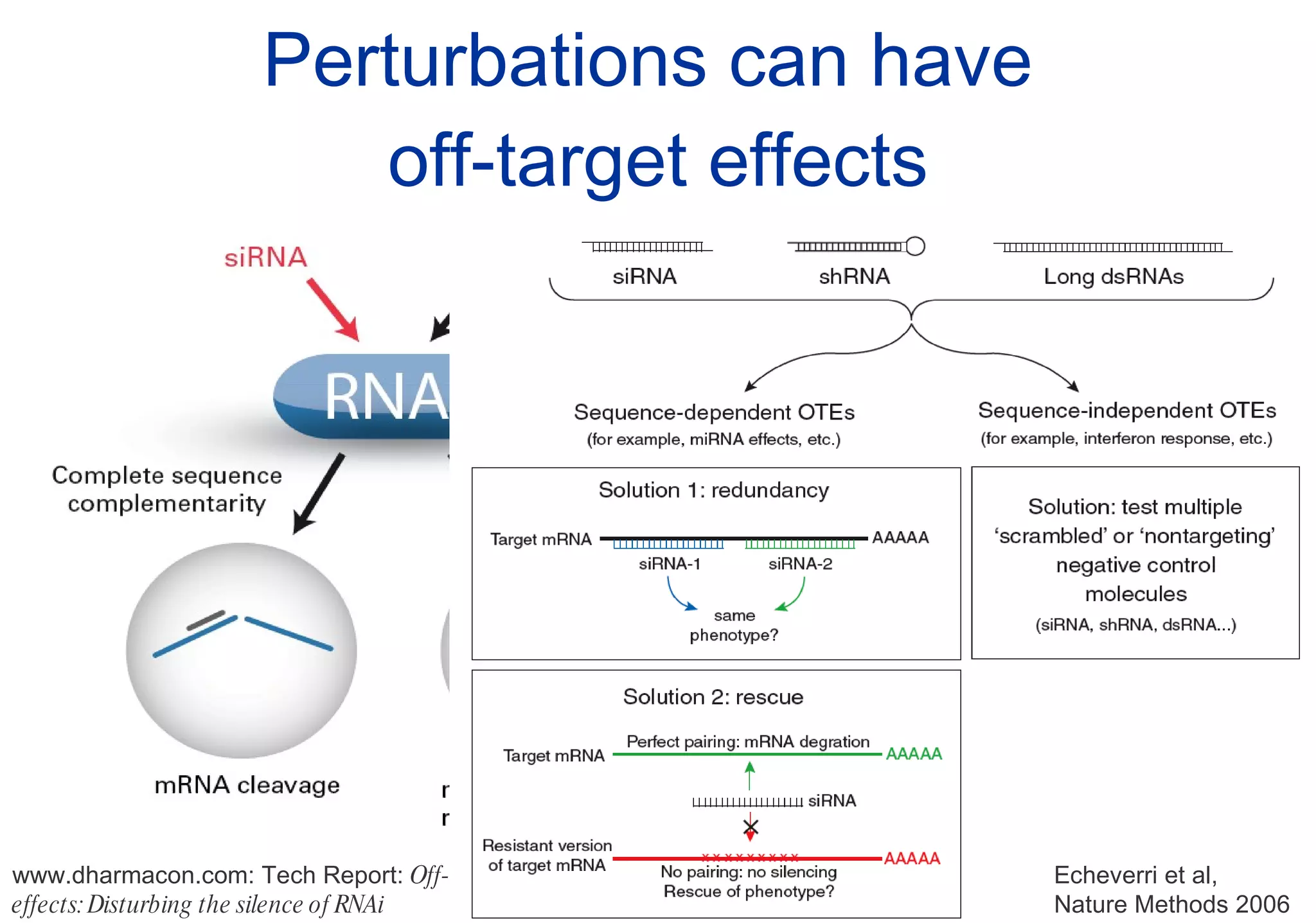
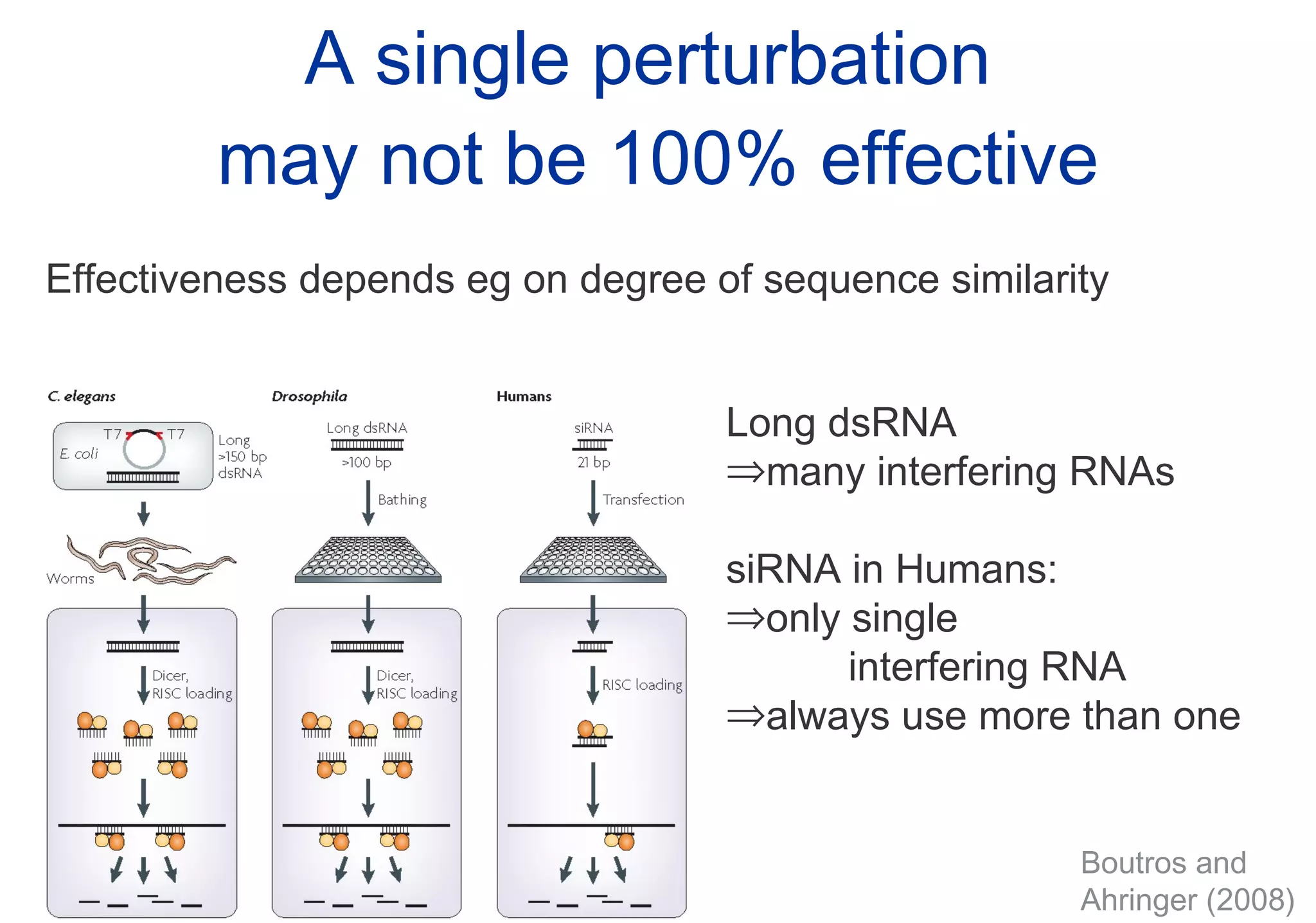
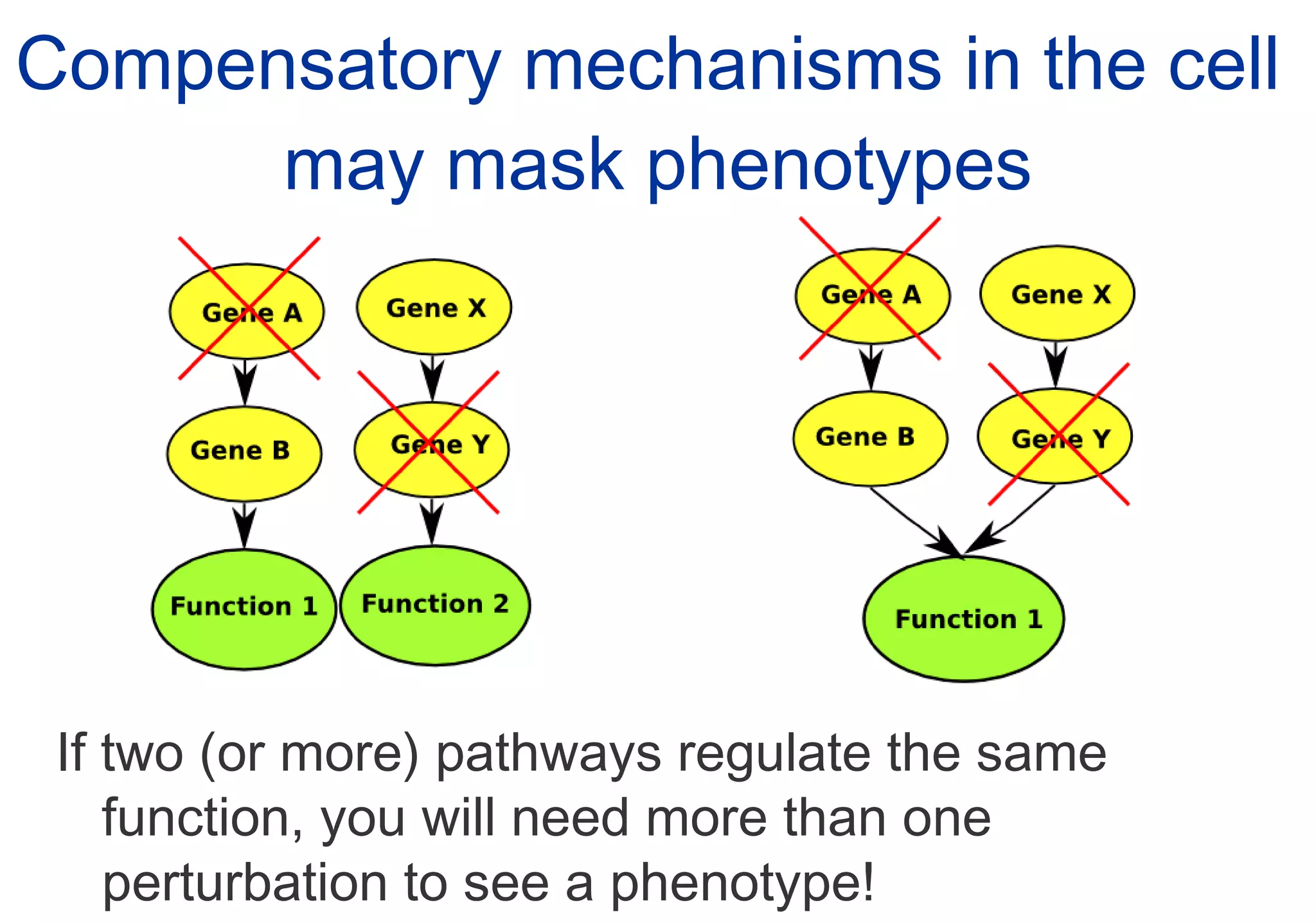
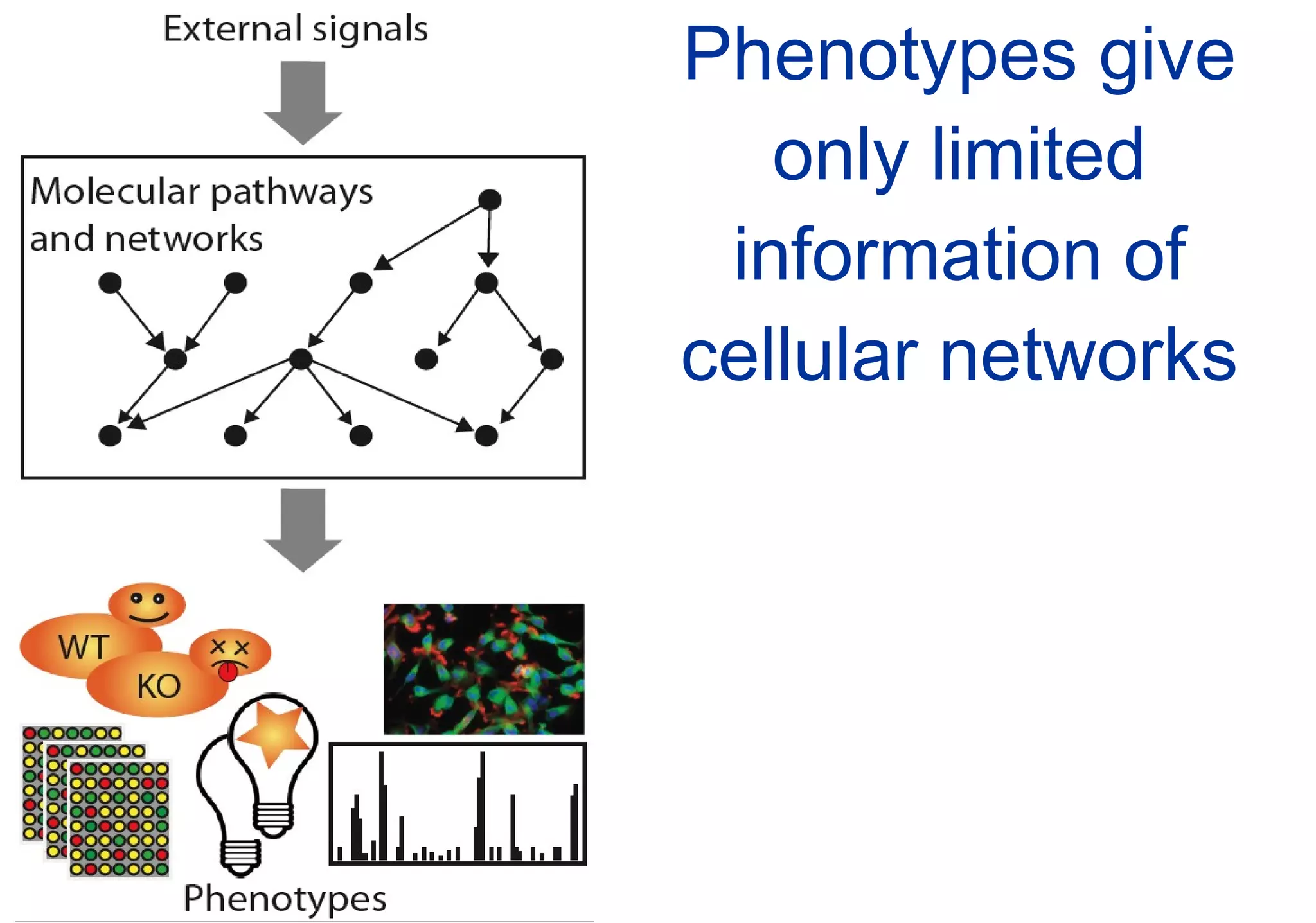
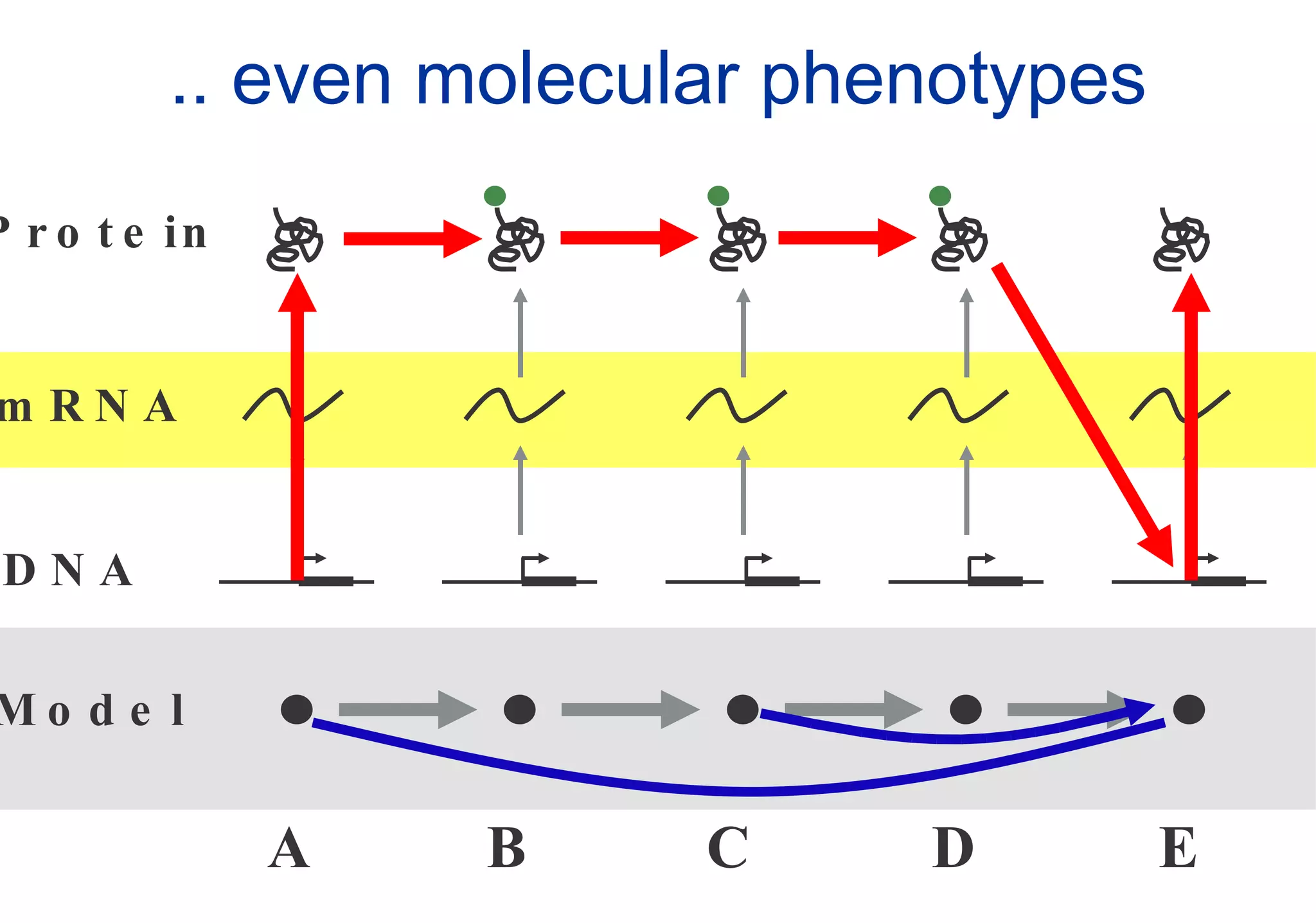
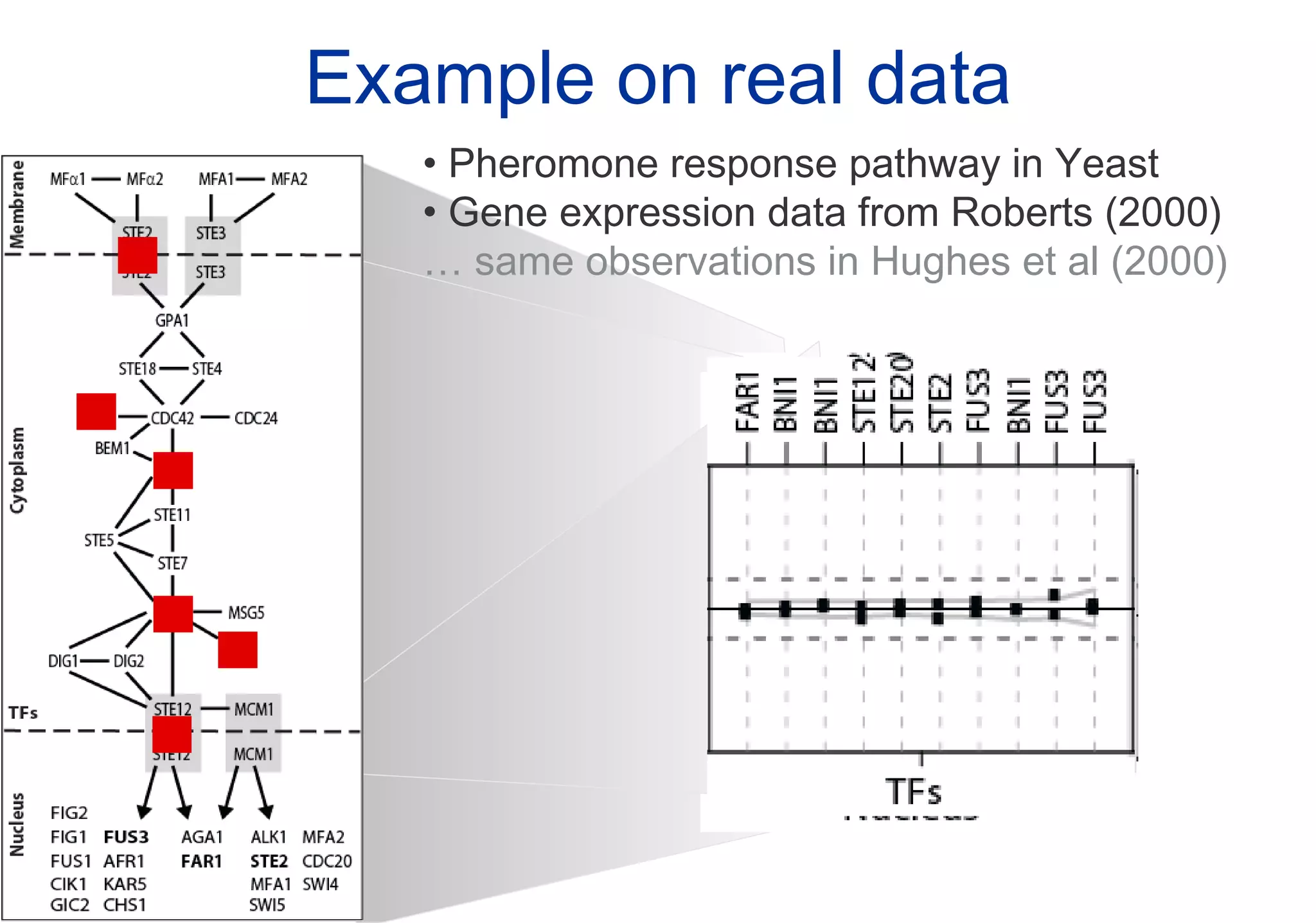
Phenotyping screens involve experimentally perturbing genes through knockouts or knockdowns and characterizing the effects on cells or organisms. Perturbations can have off-target effects or not be fully effective due to compensatory mechanisms, and phenotypes only provide limited insight into networks. Computational analysis is needed to integrate phenotype data and infer networks. Challenges include masking effects and phenotypes only revealing one aspect of cellular changes.