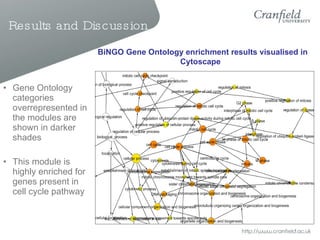
The document summarizes a thesis that aimed to identify gene expression modules in colorectal cancer using three different methods. The results conclusively identified functional gene expression modules and mapped them to known pathways. Some modules predicted tumor relapse in colorectal cancer patients and survival in breast cancer patients. The conclusions state that gene expression modules regularly occurring in colorectal cancer were identified and their functional significance was found.