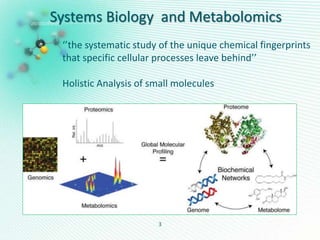
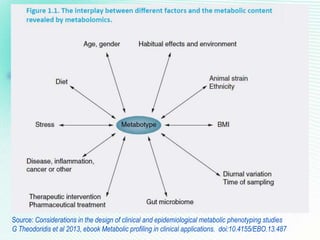
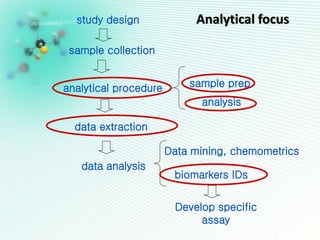
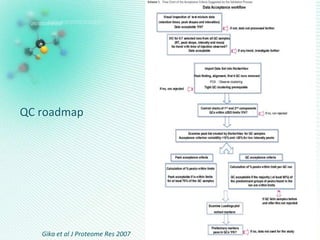
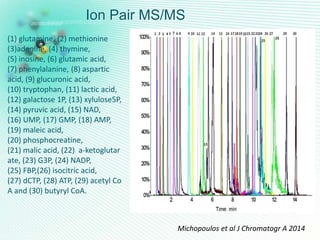
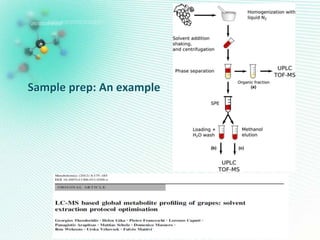
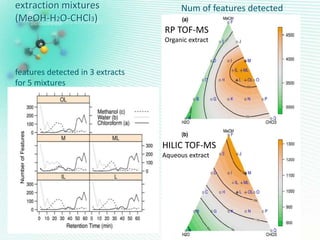
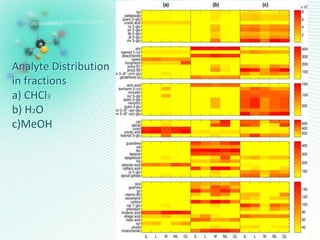
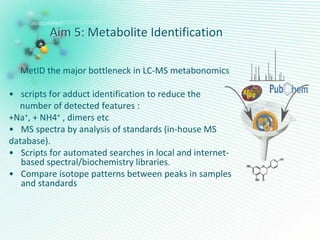
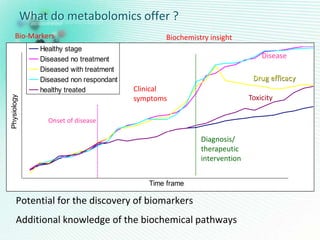
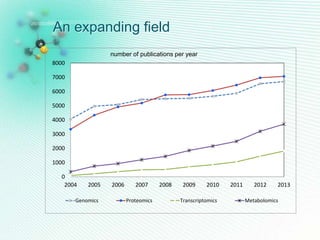
This document discusses metabolomics profiling and the challenges faced by analytical chemists. It outlines the group's work at Aristotle University on developing new analytical methods, standardizing data extraction and quality control protocols, identifying metabolites, and collaborating across disciplines. The group aims to address bottlenecks in areas like instrumentation variability, data treatment, identification, and lack of standardization. Their work seeks to advance the field and provide insights into biochemistry, biomarkers, disease, and treatment responses through holistic analysis of small molecules.