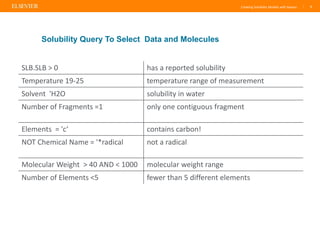
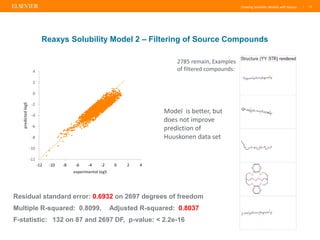
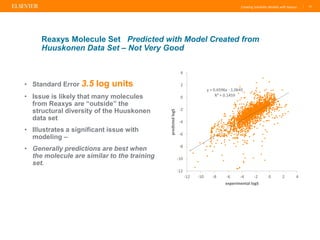
Reaxys offers a comprehensive and diverse set of solubility data that can be utilized to create predictive models, demonstrating a greater diversity than the Huuskonen data set. The document emphasizes the importance of the training set's nature and size for effective predictions, while also noting that models perform best with compounds similar to those in the training set. Additionally, Reaxys provides extensive references and conditions for measured properties, making it a valuable resource for model building in chemistry.