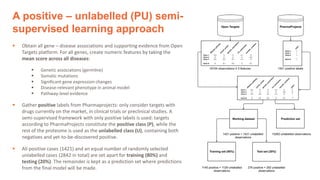
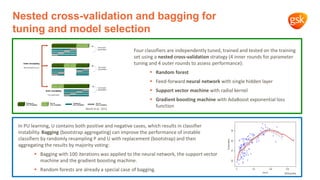
1) The document discusses using machine learning to automate drug target discovery by analyzing gene-disease association data from Open Targets.
2) A neural network classifier was able to predict therapeutic targets with 71% accuracy by analyzing features like genetic associations, somatic mutations, animal models, and pathway evidence from Open Targets.
3) The most informative features for prediction were animal models showing disease-relevant phenotypes, dysregulated gene expression in disease tissue, and genetic associations between genes and diseases.