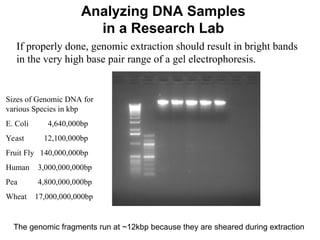
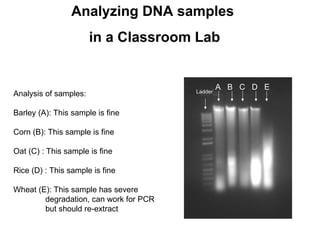
The document outlines the purpose and steps involved in DNA extraction, emphasizing its importance for further investigations like PCR and sequencing. It distinguishes between DNA extraction methods used in research labs and classroom settings, detailing procedures for lysis, precipitation, washing, and resuspension of DNA. Additionally, it assesses the quality of extracted DNA, highlighting that optimal conditions yield cleaner DNA suitable for experiments.