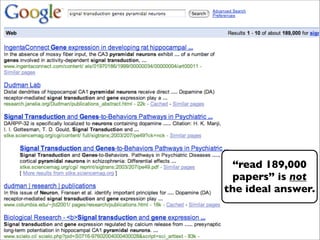
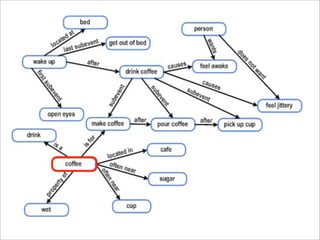
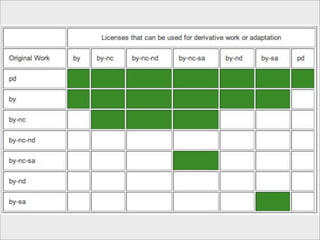
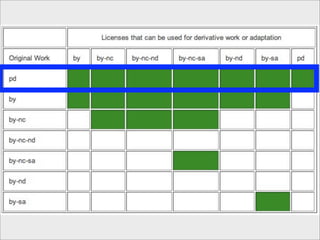
The presentation discusses the importance of knowledge sharing in the sciences, emphasizing open science and the need for accessible legal frameworks for sharing scholarly content and materials. It highlights challenges such as technical, semantic, and legal barriers to data sharing, as well as the need for interoperable formats and clear attribution practices. The document calls for a modular, standards-based approach to licensing that promotes user ease and reduces transaction costs to enable effective research collaboration.