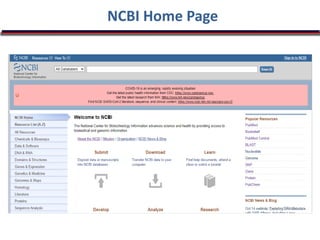
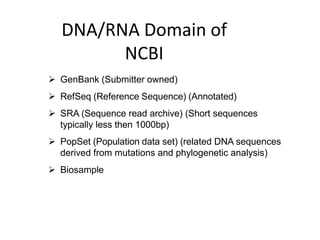
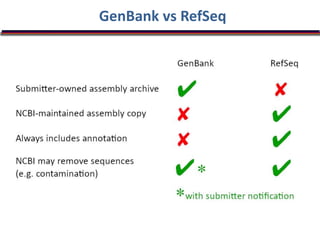
The document outlines a bioinformatics course at Abasyn University that aims to train students in analyzing genetic data, understanding protein structures, and utilizing computational tools for biological data analysis. It discusses the definition and interdisciplinary nature of bioinformatics, highlighting its reliance on computer science and mathematics to analyze biological information. Additionally, it reviews the resources provided by the National Center for Biotechnology Information (NCBI), including various databases and tools for managing biological data.