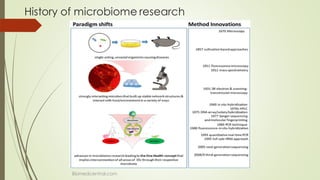
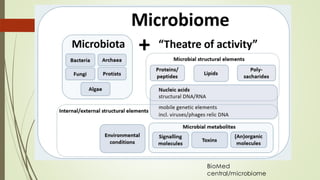
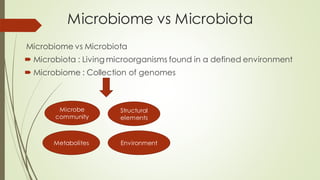
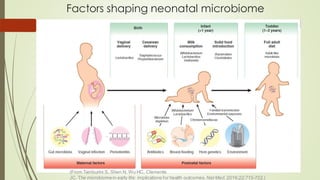
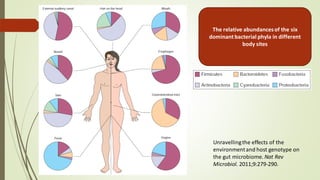
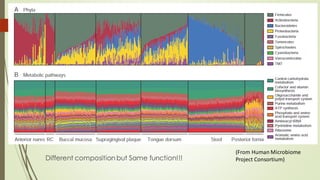
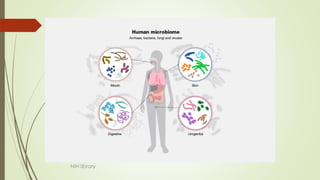
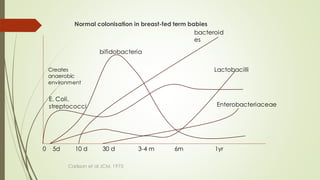
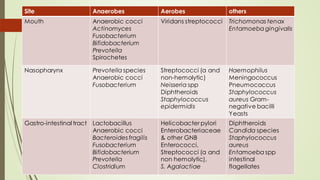
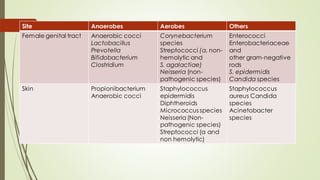
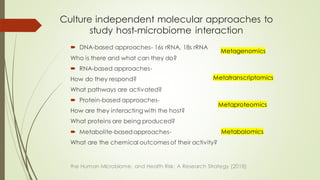
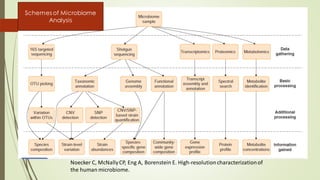
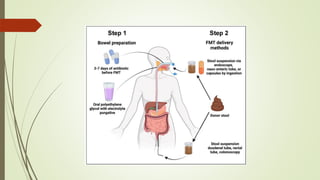
The document discusses the human microbiome, detailing its composition, roles, and factors influencing its diversity, including intrinsic and extrinsic factors such as diet and genetics. It highlights the significance of the human microbiome project in understanding the microbiota's involvement in health and disease, along with studies on fecal microbiota transplants. Various body sites and their microbial inhabitants are described, emphasizing their functions and the impact on human health.