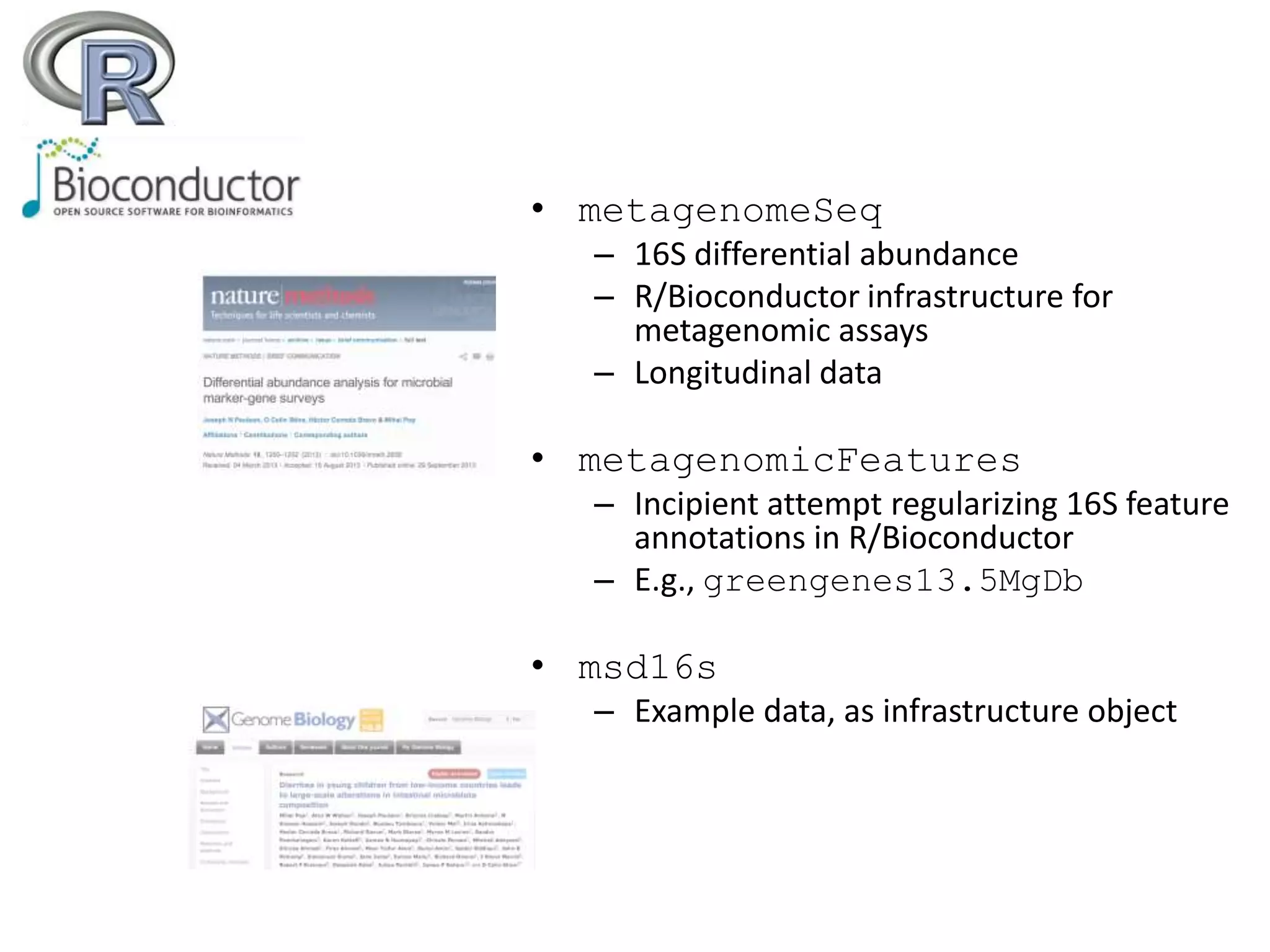
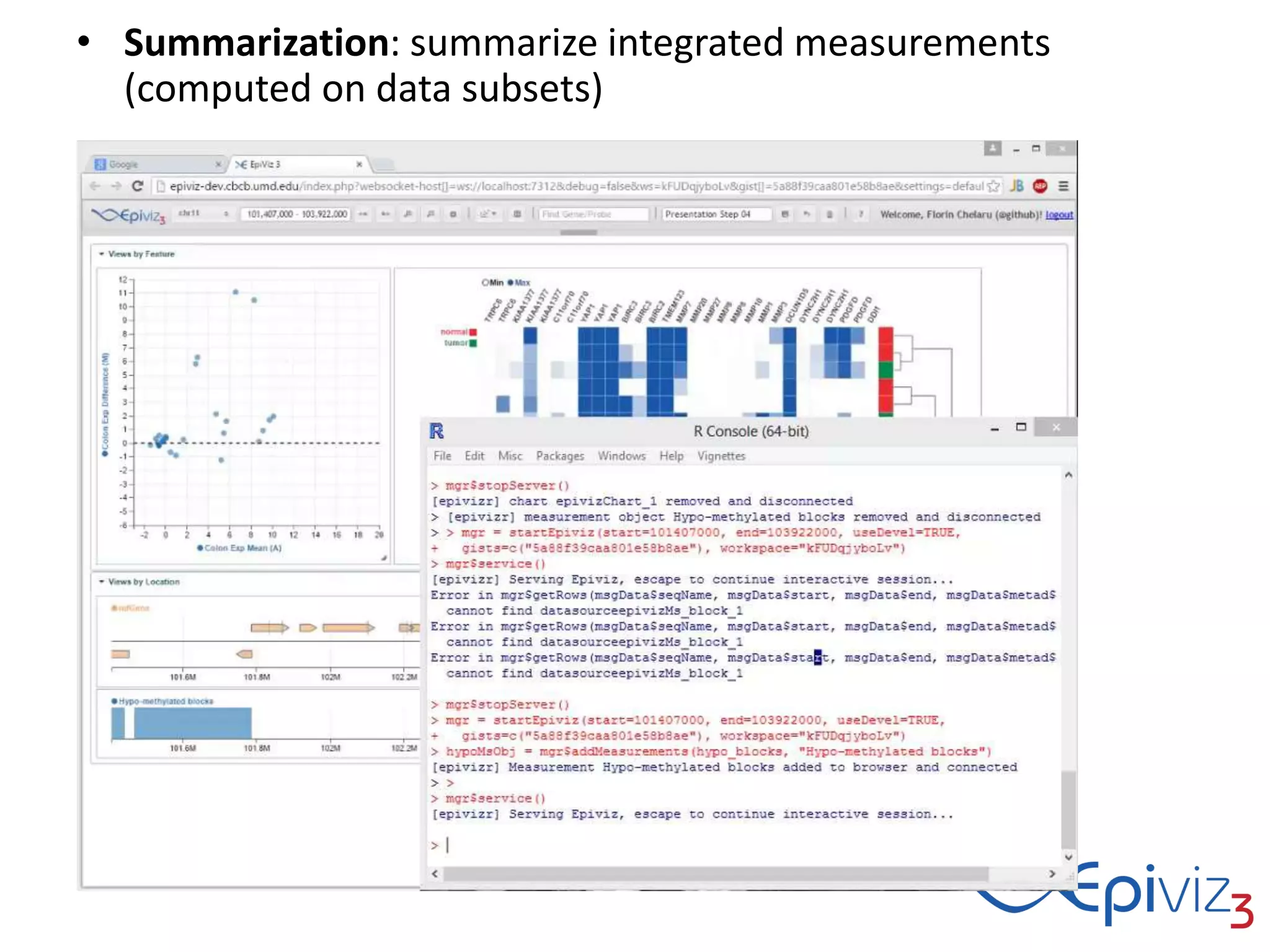
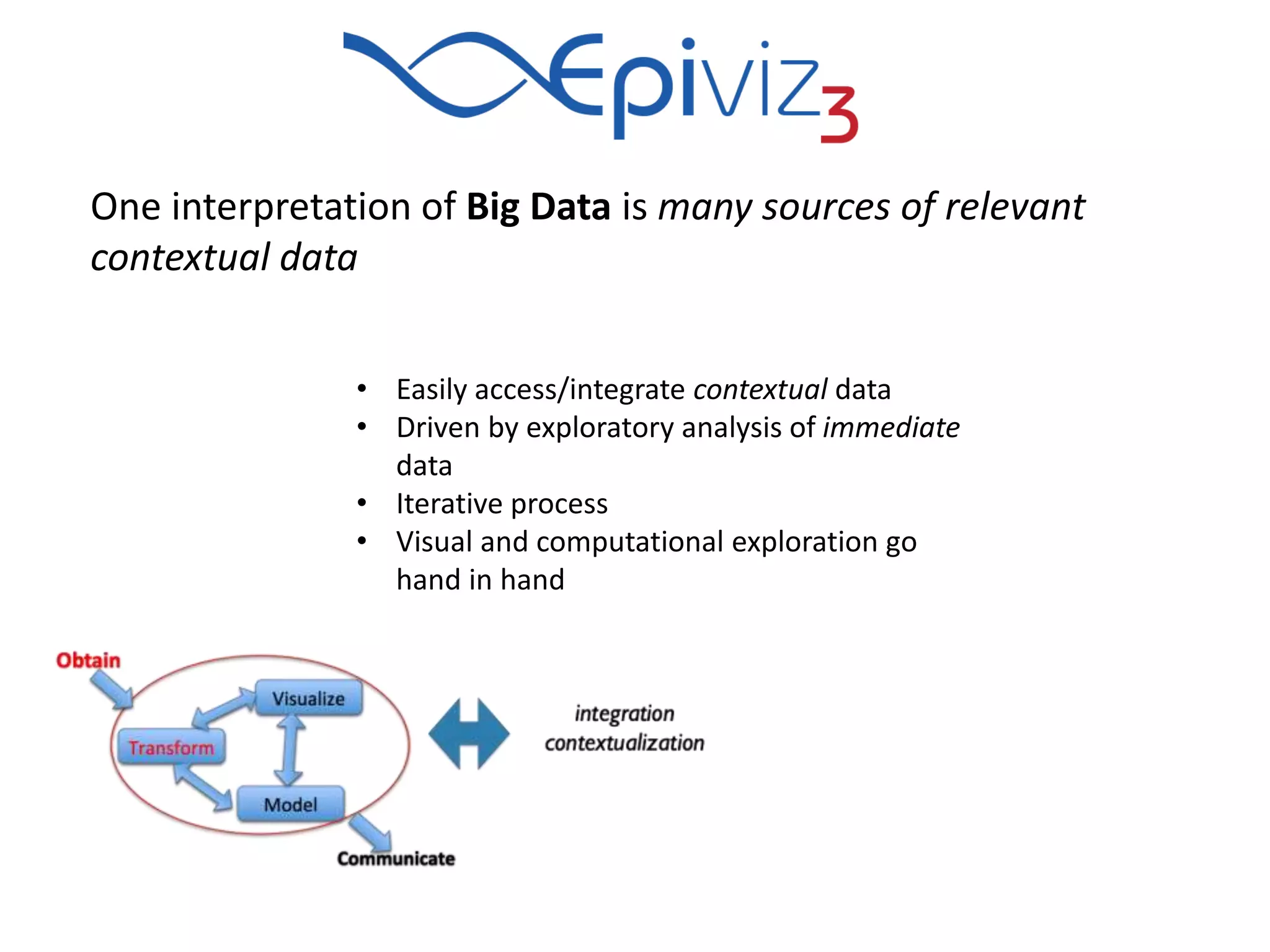
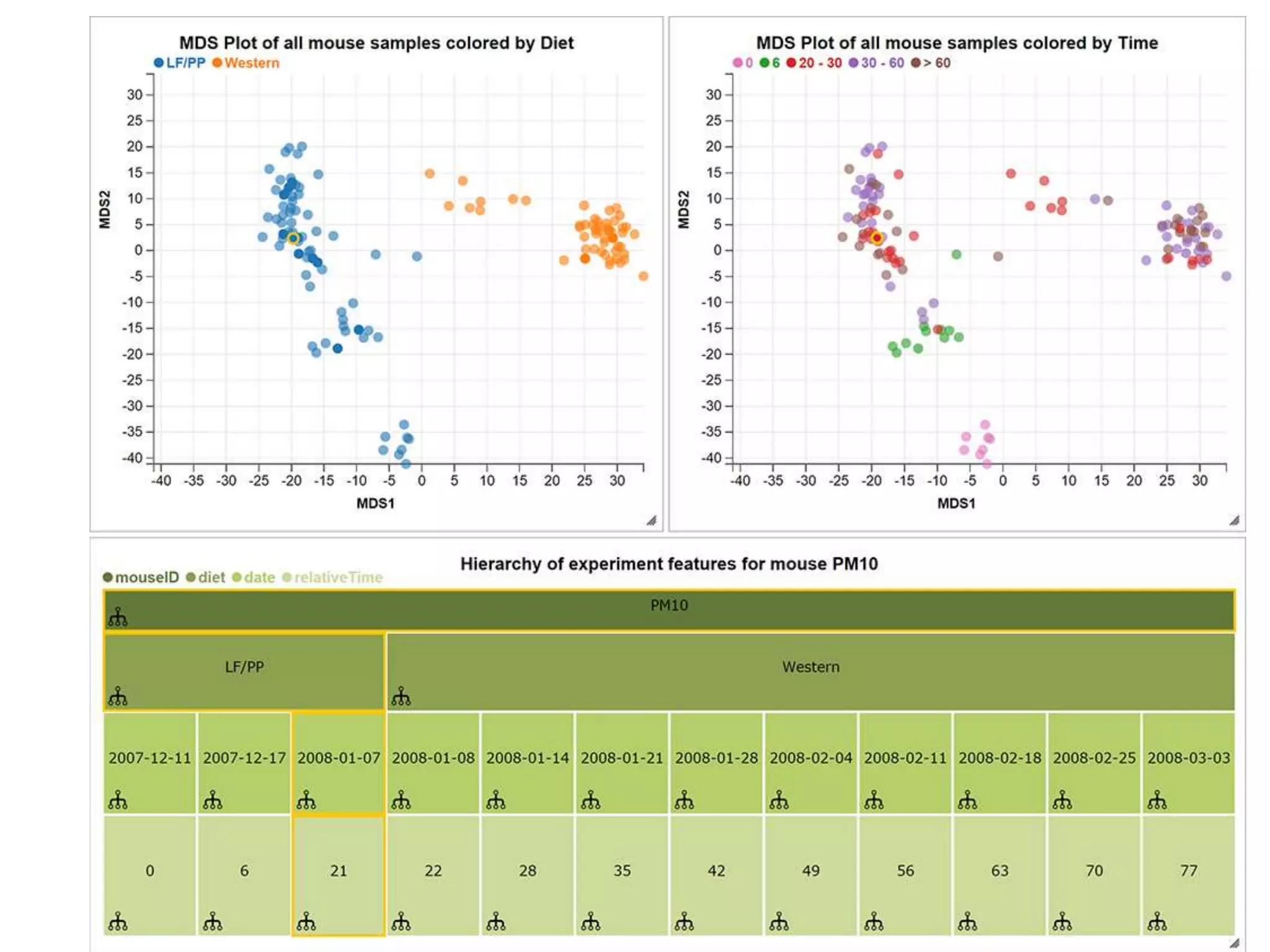
This document discusses statistical and visualization methods for analyzing metagenomic data. It introduces several R/Bioconductor packages for metagenomic analysis, including metagenomeSeq for differential abundance analysis of 16S data and metagenomicFeatures for annotating 16S features. It also describes msd16s example data. Additionally, it discusses the benefits of R/Bioconductor including infrastructure objects, documentation, and reproducibility. Finally, it introduces Metaviz, an interactive browser-based tool for exploring hierarchical metagenomic data through integration and visualization of multiple data sources.