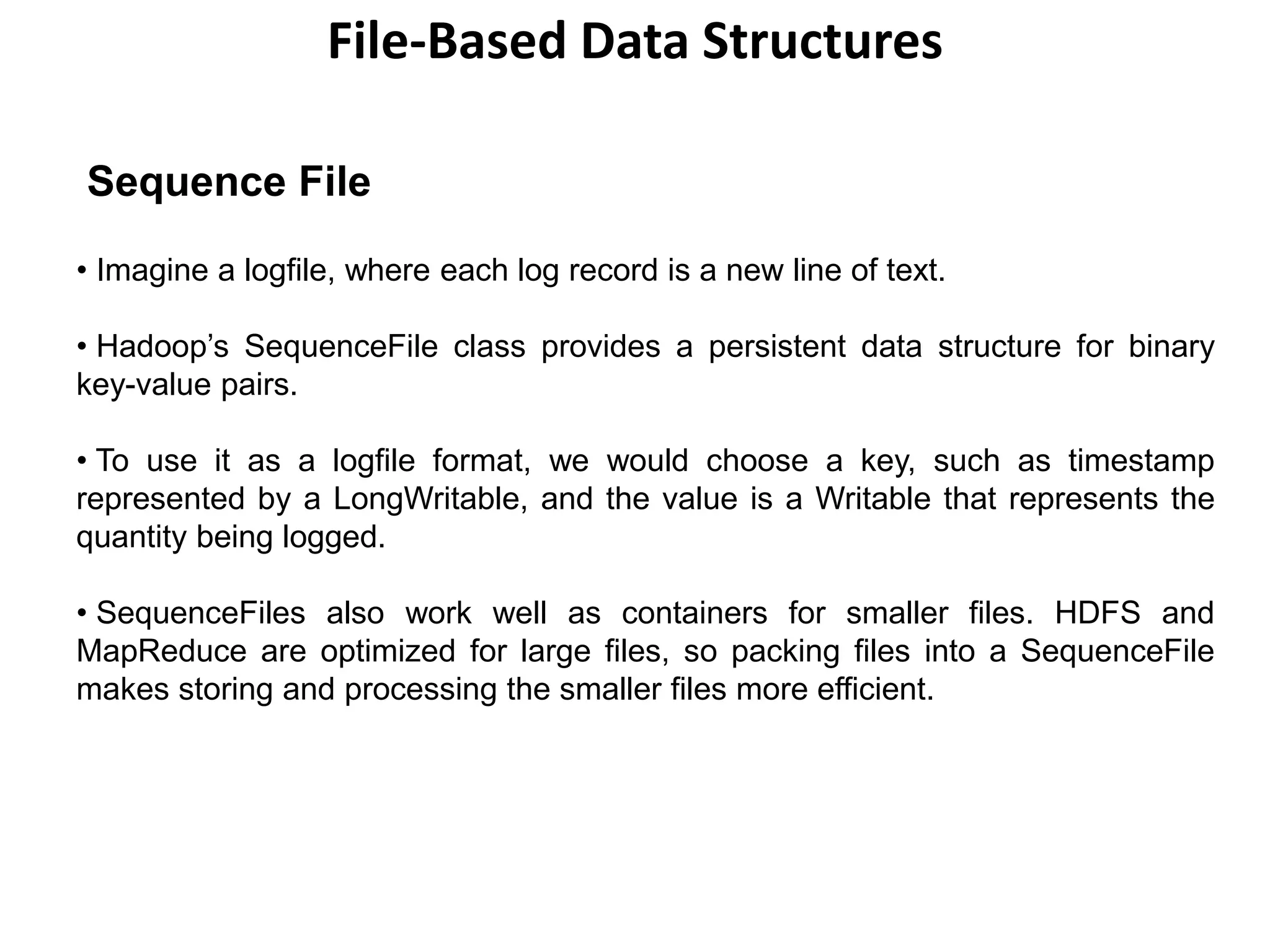
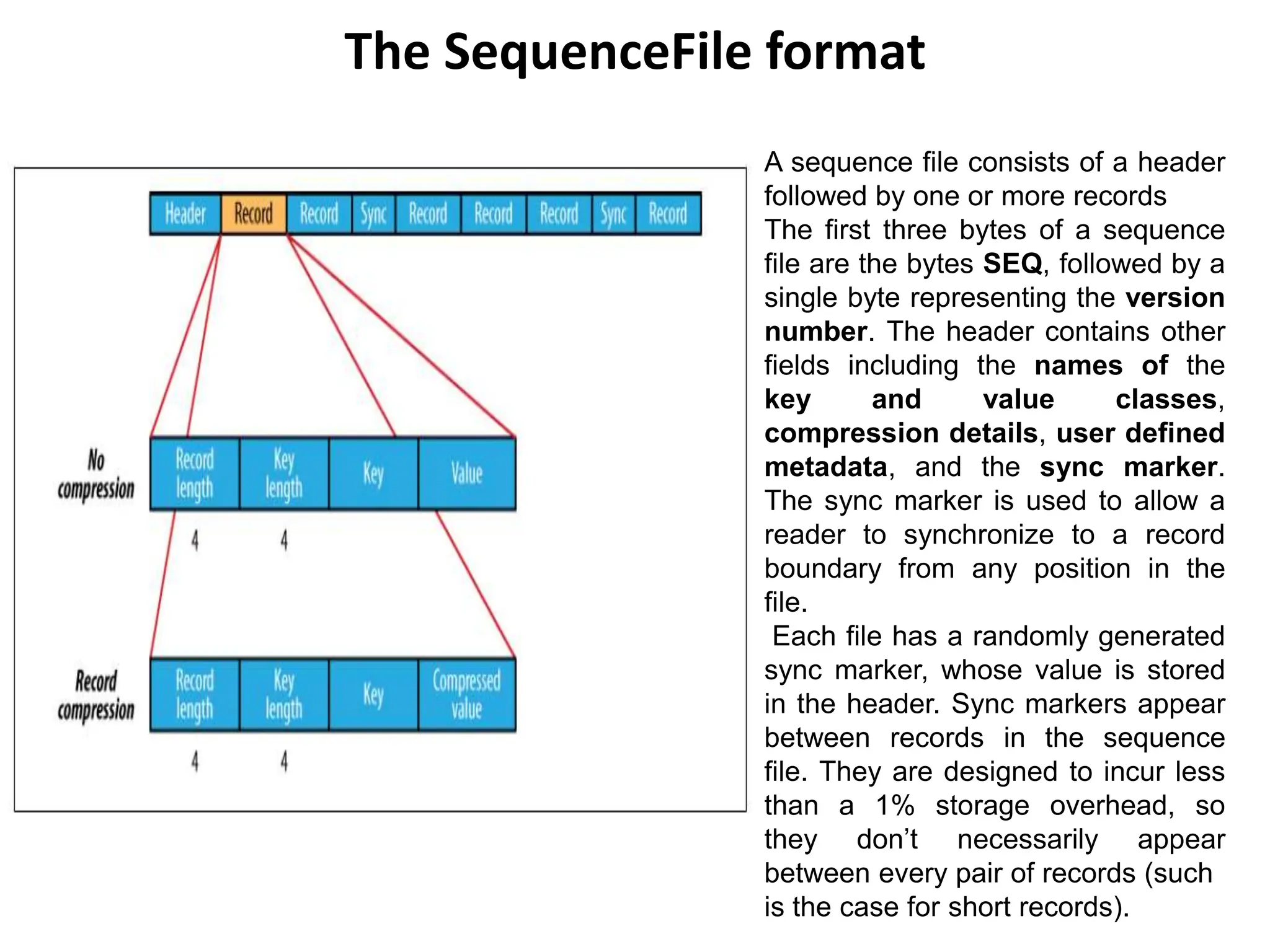
This document discusses Hadoop I/O and data integrity, serialization, compression, and file-based data structures in Hadoop.
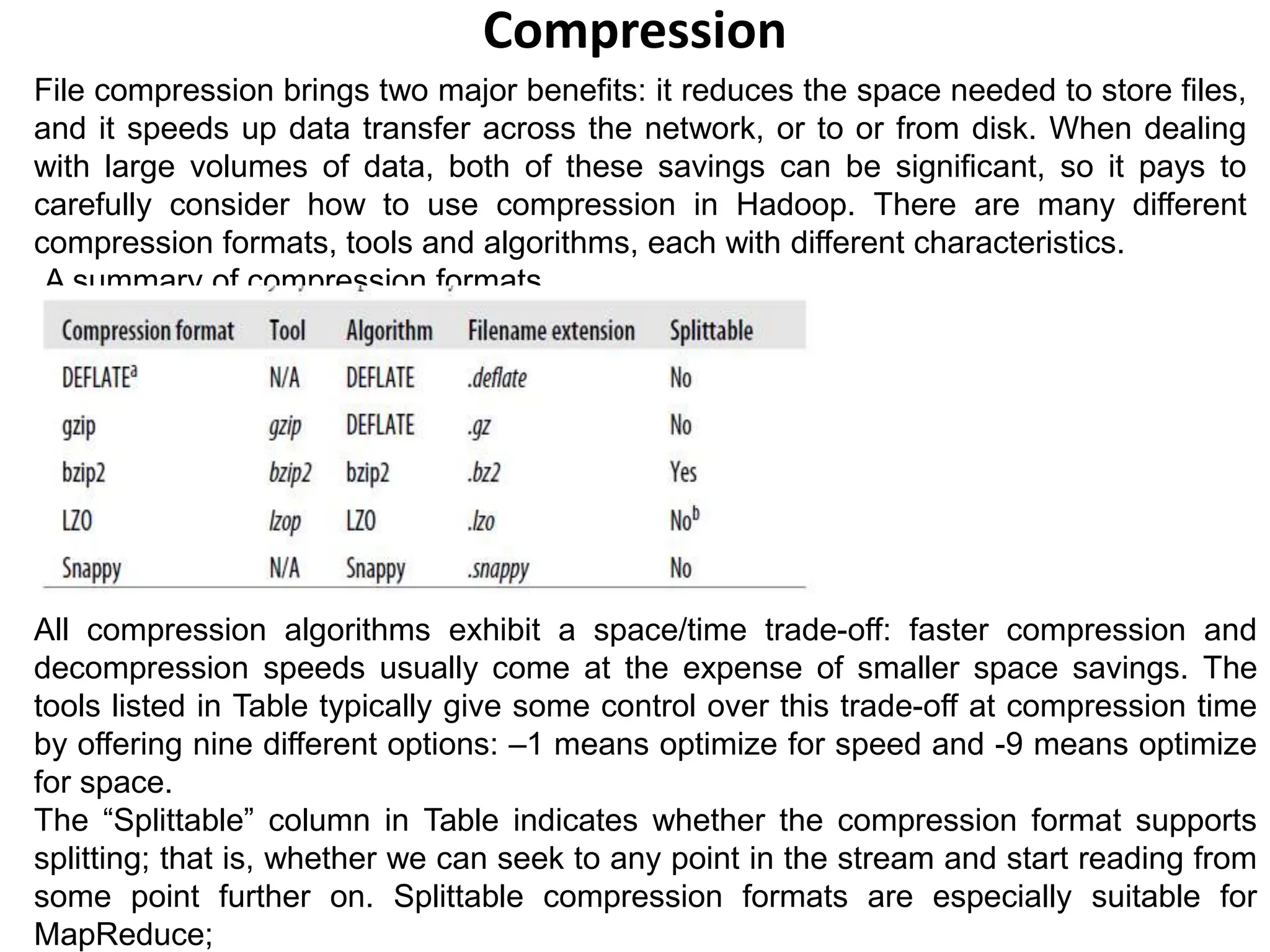
It explains that Hadoop ensures data integrity by computing and storing checksums for data blocks. When reading data, the checksum is recomputed and verified against the stored checksum to detect corruption. Compression is also discussed, including which compression formats are splittable and suitable for MapReduce.
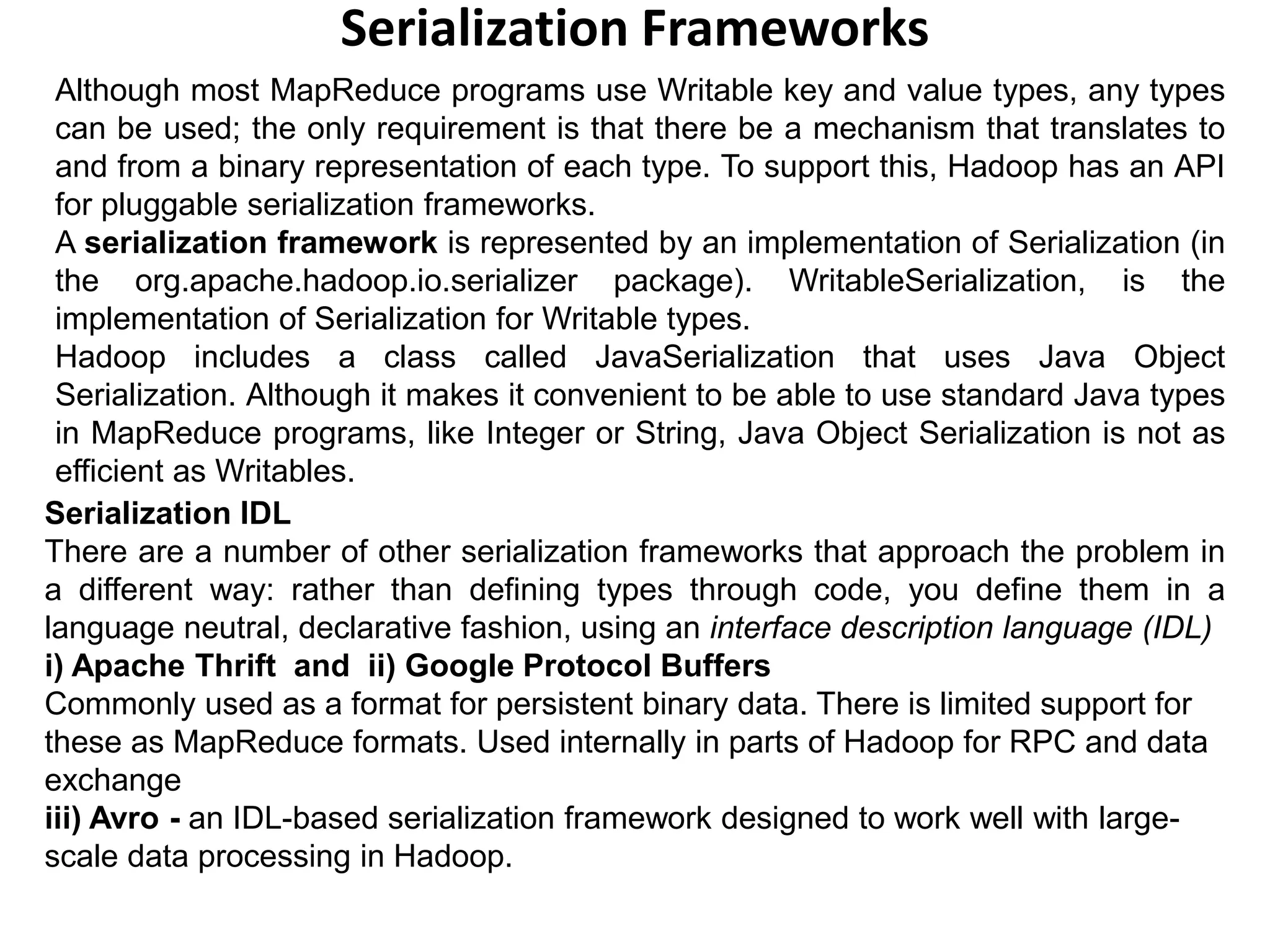
The document describes how Hadoop uses serialization to convert objects to byte streams for network transmission and storage. It introduces the Writable interface for serializable Java objects and discusses built-in Writable classes. Finally, it mentions alternative serialization frameworks that define types using an interface definition language rather than code.





![Compressing and decompressing streams with CompressionCodec
CompressionCodec has two methods that allow us to easily compress or
decompress data. To compress data being written to an output stream, use the
createOutputStream(OutputStream out) method to create a
CompressionOutputStream to which we write our uncompressed data to have it
written in compressed form to the underlying stream. To decompress data being
read from an input stream, call createInputStream(InputStream in) to obtain a
CompressionInputStream, which allows us to read uncompressed data from the
underlying stream.
API to compress data read from standard input and write it to standard output
public class StreamCompressor {
public static void main(String[] args) throws Exception {
String codecClassname = args[0];
Class<?> codecClass = Class.forName(codecClassname);
Configuration conf = new Configuration();
CompressionCodec codec = (CompressionCodec)
ReflectionUtils.newInstance(codecClass, conf);
CompressionOutputStream out = codec.createOutputStream(System.out);
IOUtils.copyBytes(System.in, out, 4096, false);
out.finish();
}
}](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-8-2048.jpg)

![Inferring CompressionCodecs using CompressionCodecFactory
A program to decompress a compressed file using a codec
inferred from the file’s extension
public class FileDecompressor {
public static void main(String[] args) throws Exception {
String uri = args[0];
Configuration conf = new Configuration();
FileSystem fs = FileSystem.get(URI.create(uri), conf);
Path inputPath = new Path(uri);
CompressionCodecFactory factory = new
CompressionCodecFactory(conf);
CompressionCodec codec = factory.getCodec(inputPath);
if (codec == null) {
System.err.println("No codec found for " + uri);
System.exit(1);
}
String outputUri =
CompressionCodecFactory.removeSuffix(uri,
codec.getDefaultExtension());
InputStream in = null;
OutputStream out = null;
try {
in = codec.createInputStream(fs.open(inputPath));
out = fs.create(new Path(outputUri));
IOUtils.copyBytes(in, out, conf);
} finally {
IOUtils.closeStream(in);
IOUtils.closeStream(out);
}
}
}
If we are reading a compressed file, we can
normally infer the codec to use by looking at its
filename extension.
A file ending in .gz can be read with
GzipCodec, and so on.
CompressionCodecFactory provides a way of
mapping a filename extension to a
CompressionCodec using its getCodec()
method, which takes a Path object for the file
in question.
Once the codec has been found, it is used to
strip off the file suffix to form the output
filename (via the removeSuffix() static method
of CompressionCodecFactory).
In this way, a file named file.gz is
decompressed to file by invoking the program
as follows:
% hadoop FileDecompressor file.gz
CompressionCodecFactory finds codecs from
a list defined by the io.compression.codecs
configuration property Each codec knows its
default filename extension, thus permitting
CompressionCodecFactory to search through
the registered codecs to find a match for a
given extension (if any).](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-10-2048.jpg)

![Using Compression in MapReduce
If our input files are compressed, they will be automatically decompressed as they are read by
MapReduce, using the filename extension to determine the codec to use. To compress the
output of a MapReduce job, in the job configuration, set the mapred.output.compress property
to true and the mapred.output.compression.codec property to the classname of the
compression codec.
public class MaxTemperatureWithCompression {
public static void main(String[] args) throws Exception {
if (args.length != 2) {
System.err.println("Usage:
MaxTemperatureWithCompression <input path> " +
"<output path>");
System.exit(-1);
}
Job job = new Job();
job.setJarByClass(MaxTemperature.class);
FileInputFormat.addInputPath(job, new Path(args[0]));
FileOutputFormat.setOutputPath(job, new Path(args[1]));
job.setOutputKeyClass(Text.class);
job.setOutputValueClass(IntWritable.class);
FileOutputFormat.setCompressOutput(job, true);
FileOutputFormat.setOutputCompressorClass(job,
GzipCodec.class);
job.setMapperClass(MaxTemperatureMapper.class);
job.setCombinerClass(MaxTemperatureReducer.class);
job.setReducerClass(MaxTemperatureReducer.class);
System.exit(job.waitForCompletion(true) ? 0 : 1);
} }
% hadoop
MaxTemperatureWithCompression
input/ncdc/sample.txt.gz
output
Each part of the final output is
compressed; in this case, there is a single
part:
% gunzip -c output/part-r-00000.gz
1949 111
1950 22](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-12-2048.jpg)


![The Writable Interface
The Writable interface defines two methods: one for writing its state to a
DataOutput binary stream, and one for reading its state from a DataInput binary
stream:
Let’s look at a particular Writable to see what we can do with it. We
will use IntWritable, a wrapper for a Java int. We can create one
and set its value using the set() method:
IntWritable writable = new IntWritable();
writable.set(163);
Equivalently, we can use the constructor that takes the integer
value:
IntWritable writable = new IntWritable(163);
To examine the serialized form of the IntWritable, we write a small
helper method that wraps a java.io.ByteArrayOutputStream in a
java.io.DataOutputStream (to capture the bytes in the serialized
stream:
public static byte[] serialize(Writable writable) throws IOException {
ByteArrayOutputStream out = new ByteArrayOutputStream();
DataOutputStream dataOut = new DataOutputStream(out);
writable.write(dataOut);
dataOut.close();
return out.toByteArray();
}](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-15-2048.jpg)
![The Writable Interface
An integer is written using four bytes (as we see using JUnit 4 assertions):
byte[] bytes = serialize(writable);
assertThat(bytes.length, is(4));
The bytes are written in big-endian order (so the most significant byte is written
to the stream first, this is dictated by the java.io.DataOutput interface), and we
can see their hexadecimal representation by using a method on Hadoop’s
StringUtils:
assertThat(StringUtils.byteToHexString(bytes), is("000000a3"));
Let’s try deserialization. Again, we create a helper method to read a Writable
object from a byte array:
public static byte[] deserialize(Writable writable, byte[] bytes)
throws IOException {
ByteArrayInputStream in = new ByteArrayInputStream(bytes);
DataInputStream dataIn = new DataInputStream(in);
writable.readFields(dataIn);
dataIn.close();
return bytes;
}](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-16-2048.jpg)







![In-memory serialization and deserialization
Let’s write a Java program to read and write Avro data to and from streams. We’ll
start with a simple Avro schema for representing a pair of strings as a record:
{ "type": "record",
"name": "StringPair",
"doc": "A pair of strings.",
"fields": [
{"name": "left", "type": "string"},
{"name": "right", "type": "string"} ] }
If this schema is saved in a file on the classpath called StringPair.avsc (.avsc is the
conventional extension for an Avro schema), then we can load it using the following
two lines of code:
Schema.Parser parser = new Schema.Parser();
Schema schema =
parser.parse(getClass().getResourceAsStream("StringPair.avsc"));
We can create an instance of an Avro record using the generic API as follows:
GenericRecord datum = new GenericData.Record(schema);
datum.put("left", "L");
datum.put("right", "R");](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-25-2048.jpg)


![A C program for reading Avro record pairs from a data file
#include <avro.h>
#include <stdio.h>
#include <stdlib.h>
int main(int argc, char *argv[]) {
if (argc != 2) {
fprintf(stderr, "Usage: dump_pairs <data_file>n");
exit(EXIT_FAILURE);
}
const char *avrofile = argv[1];
avro_schema_error_t error;
avro_file_reader_t filereader;
avro_datum_t pair;
avro_datum_t left;
avro_datum_t right;
int rval;
char *p;
avro_file_reader(avrofile, &filereader);
while (1) {
rval = avro_file_reader_read(filereader, NULL, &pair);
if (rval) break;
if (avro_record_get(pair, "left", &left) == 0) {
avro_string_get(left, &p);
fprintf(stdout, "%s,", p); }
if (avro_record_get(pair, "right", &right) == 0) {
avro_string_get(right, &p);
fprintf(stdout, "%sn", p); } }
avro_file_reader_close(filereader);
return 0; }
The core of the program does three things:
1. opens a file reader of type avro_file_reader_t
by calling Avro’s
avro_file_reader function
2. reads Avro data from the file reader with the
avro_file_reader_read function in a
while loop until there are no pairs left (as
determined by the return value rval), and
3. closes the file reader with
avro_file_reader_close.
Running the program using the output of
the Python program prints the original
input:
% ./dump_pairs pairs.avro
a,1
c,2
b,3
b,2
We have successfully exchanged complex
data between two Avro implementations.
Interoperability](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-28-2048.jpg)
![Interoperability
A Python program for writing Avro record pairs to a data file
import os
import string
import sys
from avro import schema
from avro import io
from avro import datafile
if __name__ == '__main__':
if len(sys.argv) != 2:
sys.exit('Usage: %s <data_file>' % sys.argv[0])
avro_file = sys.argv[1]
writer = open(avro_file, 'wb')
datum_writer = io.DatumWriter()
schema_object = schema.parse("""
{ "type": "record",
"name": "StringPair",
"doc": "A pair of strings.",
"fields": [
{"name": "left", "type": "string"},
{"name": "right", "type": "string"}
]
}""")
dfw = datafile.DataFileWriter(writer, datum_writer,
schema_object)
for line in sys.stdin.readlines():
(left, right) = string.split(line.strip(), ',')
dfw.append({'left':left, 'right':right});
dfw.close()
The program reads comma-separated strings
from standard input and writes them as StringPair
records to an Avro data file. Like the Java code
for writing a data file, we create a DatumWriter
and a DataFileWriter object. Notice that we have
embedded the Avro schema in the code,
although we could equally well have read it from
a file.
Python represents Avro records as dictionaries;
each line that is read from standard in is turned
into a dict object and appended to the
DataFileWriter.
Before we can run the program, we need to install Avro
for Python:
% easy_install avro
To run the program, we specify the name of the file to
write output to (pairs.avro) and
send input pairs over standard in, marking the end of file
by typing Control-D:
% python avro/src/main/py/write_pairs.py pairs.avro
a,1
c,2
b,3
b,2
^D](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-29-2048.jpg)
![Writing a SequenceFile
Program to write some key-value pairs to a SequenceFile
private static final String[] DATA = {
"One, two, buckle my shoe",
"Three, four, shut the door",
"Five, six, pick up sticks",
"Seven, eight, lay them straight",
"Nine, ten, a big fat hen"
};
public static void main(String[] args) throws IOException {
String uri = args[0];
Configuration conf = new Configuration();
FileSystem fs = FileSystem.get(URI.create(uri), conf);
Path path = new Path(uri);
IntWritable key = new IntWritable();
Text value = new Text();
SequenceFile.Writer writer = null;
try {
writer = SequenceFile.createWriter(fs, conf, path,
key.getClass(), value.getClass());
for (int i = 0; i < 100; i++) {
key.set(100 - i);
value.set(DATA[i % DATA.length]);
System.out.printf("[%s]t%st%sn", writer.getLength(), key,
value);
writer.append(key, value); }
} finally {
IOUtils.closeStream(writer);
}}}
% hadoop SequenceFileWriteDemo
numbers.seq
[128] 100 One, two, buckle my shoe
[173] 99 Three, four, shut the door
[220] 98 Five, six, pick up sticks
[264] 97 Seven, eight, lay them straight
[314] 96 Nine, ten, a big fat hen
[359] 95 One, two, buckle my shoe
[404] 94 Three, four, shut the door
[451] 93 Five, six, pick up sticks
[495] 92 Seven, eight, lay them straight
[545] 91 Nine, ten, a big fat hen
...
[1976] 60 One, two, buckle my shoe
[2021] 59 Three, four, shut the door
[2088] 58 Five, six, pick up sticks
[2132] 57 Seven, eight, lay them straight
[2182] 56 Nine, ten, a big fat hen
...
[4557] 5 One, two, buckle my shoe
[4602] 4 Three, four, shut the door
[4649] 3 Five, six, pick up sticks
[4693] 2 Seven, eight, lay them straight
[4743] 1 Nine, ten, a big fat hen](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-32-2048.jpg)


![Reading a SequenceFile
public class SequenceFileReadDemo {
public static void main(String[] args) throws IOException {
String uri = args[0];
Configuration conf = new Configuration();
FileSystem fs = FileSystem.get(URI.create(uri), conf);
Path path = new Path(uri);
SequenceFile.Reader reader = null;
try {
reader = new SequenceFile.Reader(fs, path, conf);
Writable key = (Writable)
ReflectionUtils.newInstance(reader.getKeyClass(), conf);
Writable value = (Writable)
ReflectionUtils.newInstance(reader.getValueClass(), conf);
long position = reader.getPosition();
while (reader.next(key, value)) {
String syncSeen = reader.syncSeen() ? "*" : "";
System.out.printf("[%s%s]t%st%sn", position, syncSeen, key, value);
position = reader.getPosition(); // beginning of next record
}
} finally {
IOUtils.closeStream(reader);
}
}
}](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-35-2048.jpg)
![% hadoop SequenceFileReadDemo numbers.seq
[128] 100 One, two, buckle my shoe
[173] 99 Three, four, shut the door
[220] 98 Five, six, pick up sticks
[264] 97 Seven, eight, lay them straight
[314] 96 Nine, ten, a big fat hen
[359] 95 One, two, buckle my shoe
[404] 94 Three, four, shut the door
[451] 93 Five, six, pick up sticks
[495] 92 Seven, eight, lay them straight
[545] 91 Nine, ten, a big fat hen
[590] 90 One, two, buckle my shoe
...
[1976] 60 One, two, buckle my shoe
[2021*] 59 Three, four, shut the door
[2088] 58 Five, six, pick up sticks
[2132] 57 Seven, eight, lay them straight
[2182] 56 Nine, ten, a big fat hen
...
[4557] 5 One, two, buckle my shoe
[4602] 4 Three, four, shut the door
[4649] 3 Five, six, pick up sticks
[4693] 2 Seven, eight, lay them straight
[4743] 1 Nine, ten, a big fat hen](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-36-2048.jpg)


![Writing a MapFile
public class MapFileWriteDemo {
private static final String[] DATA = {
"One, two, buckle my shoe",
"Three, four, shut the door",
"Five, six, pick up sticks",
"Seven, eight, lay them straight",
"Nine, ten, a big fat hen"
};
public static void main(String[] args) throws IOException {
String uri = args[0];
Configuration conf = new Configuration();
FileSystem fs = FileSystem.get(URI.create(uri), conf);
IntWritable key = new IntWritable();
Text value = new Text();
MapFile.Writer writer = null;
try {
writer = new MapFile.Writer(conf, fs, uri,
key.getClass(), value.getClass());
for (int i = 0; i < 1024; i++) {
key.set(i + 1);
value.set(DATA[i % DATA.length]);
writer.append(key, value);
}
} finally {
IOUtils.closeStream(writer); }}}](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-40-2048.jpg)



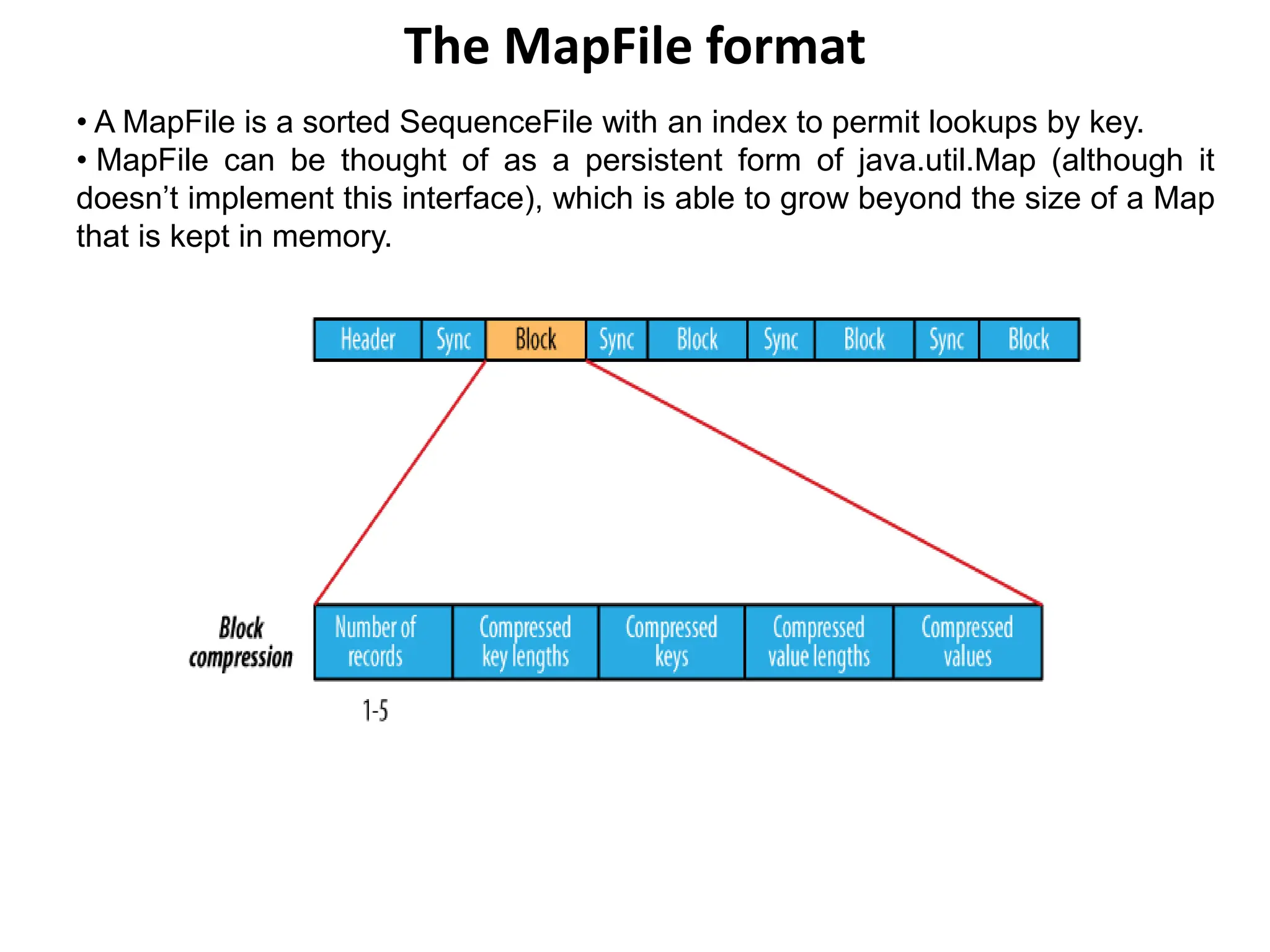
![Converting a SequenceFile to a MapFile
One way of looking at a MapFile is as an indexed and sorted SequenceFile. So it’s
quite natural to want to be able to convert a SequenceFile into a MapFile.
public class MapFileFixer {
public static void main(String[] args) throws Exception {
String mapUri = args[0];
Configuration conf = new Configuration();
FileSystem fs = FileSystem.get(URI.create(mapUri), conf);
Path map = new Path(mapUri);
Path mapData = new Path(map, MapFile.DATA_FILE_NAME);
// Get key and value types from data sequence file
SequenceFile.Reader reader = new SequenceFile.Reader(fs, mapData, conf);
Class keyClass = reader.getKeyClass();
Class valueClass = reader.getValueClass();
reader.close();
// Create the map file index file
long entries = MapFile.fix(fs, map, keyClass, valueClass, false, conf);
System.out.printf("Created MapFile %s with %d entriesn", map, entries);
}
}](https://image.slidesharecdn.com/hadoooio-240201143110-5634f46a/75/HadoooIO-ppt-44-2048.jpg)
