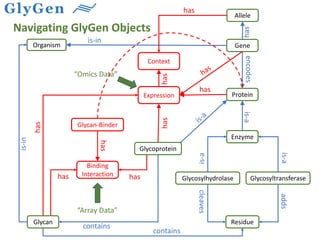
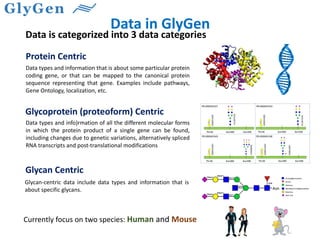
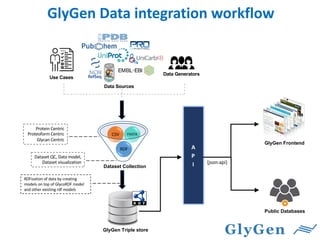
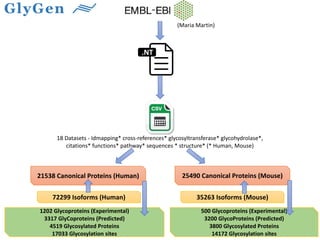
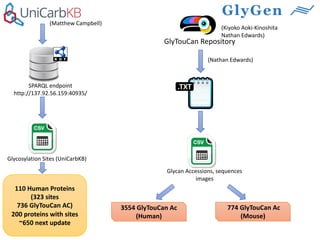
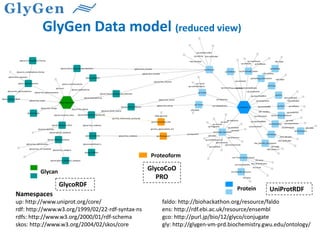
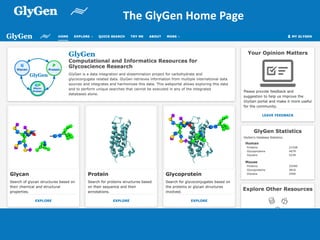
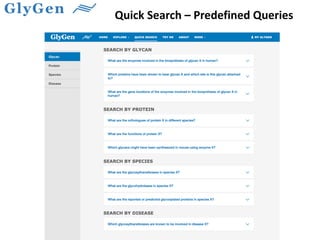
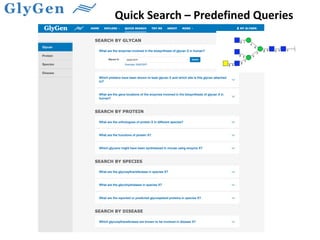
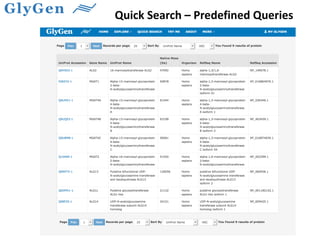
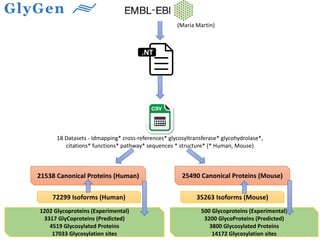
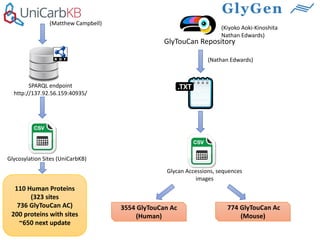
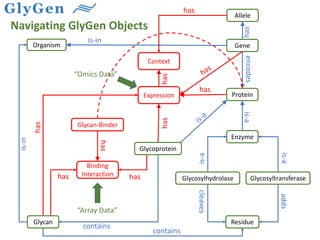
GlySpace is a collaborative project between several universities to create GlyGen, a glycoinformatics resource. GlyGen will integrate various glycan, protein, and glycoprotein data from public databases. It will have a SPARQL endpoint and web interface to allow searching and exploring relationships between glycans, proteins, enzymes and other glycoinformatics data. The goal is to facilitate research in glycobiology and disease.