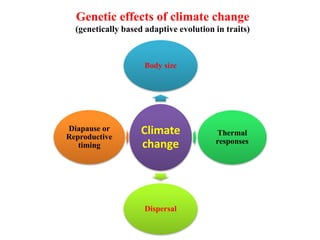
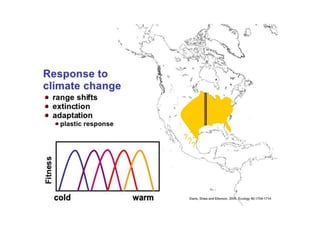
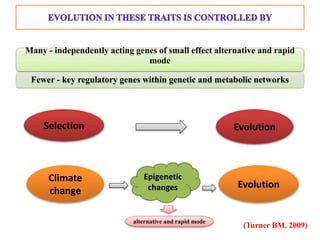
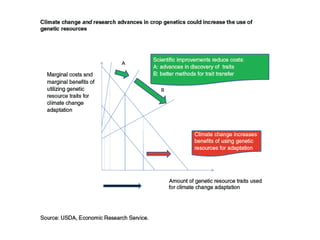
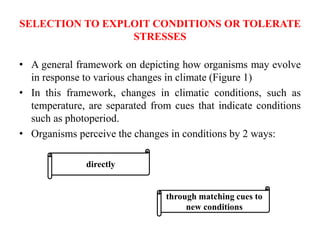
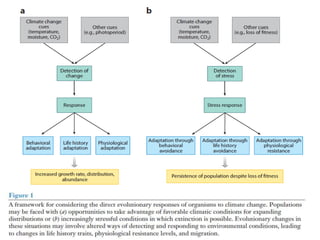
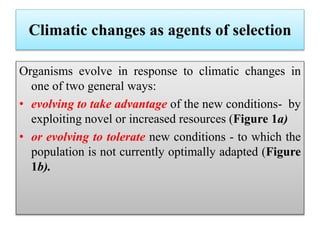
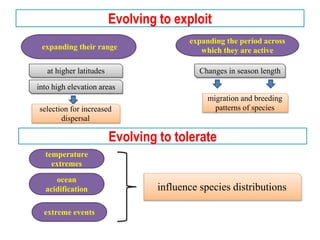
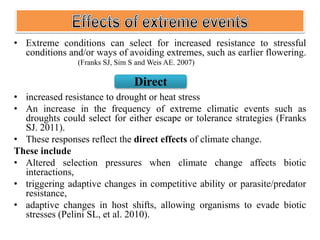
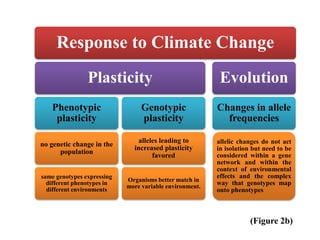
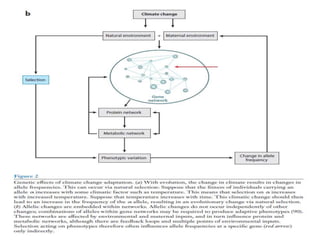
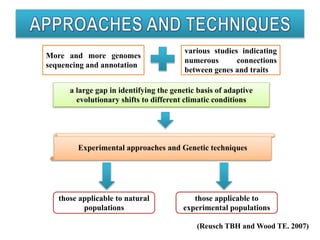
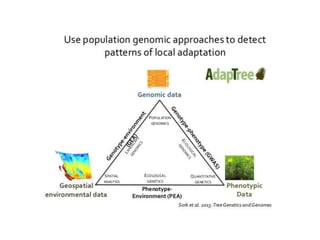
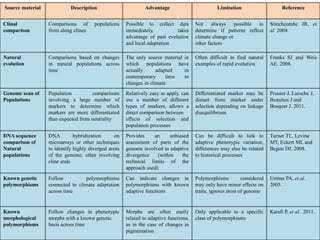
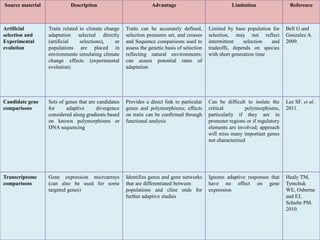
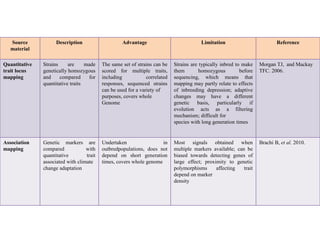
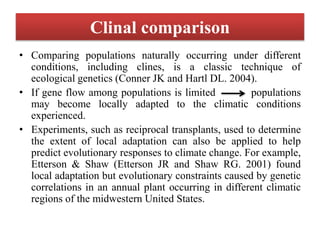
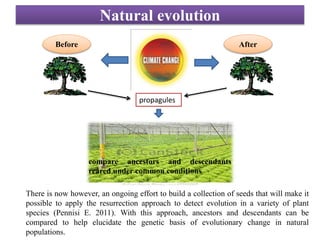
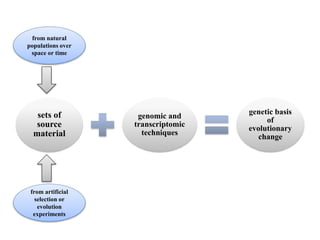
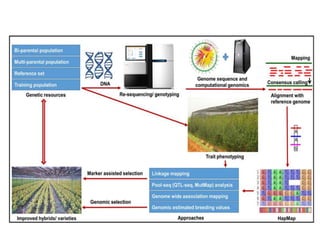
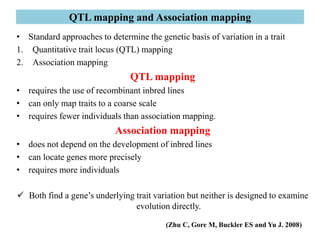
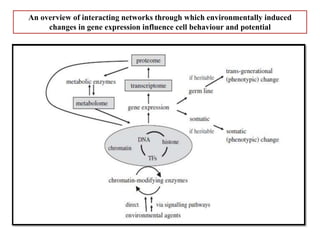
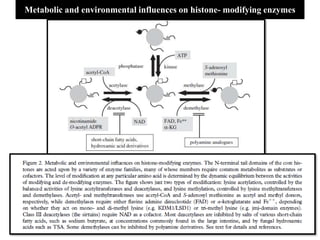
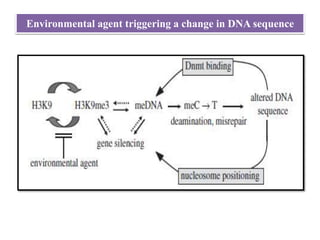
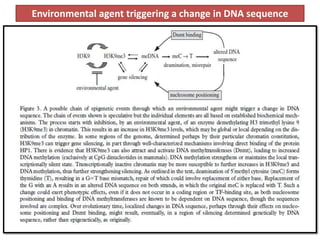
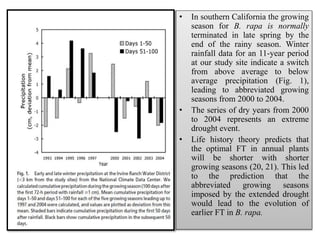
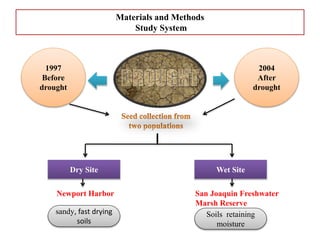
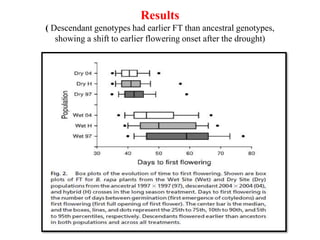
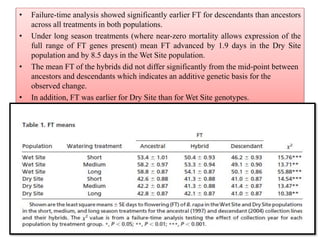
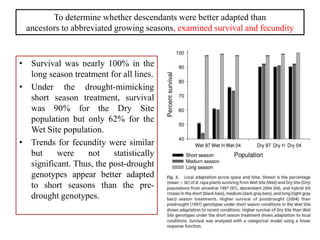
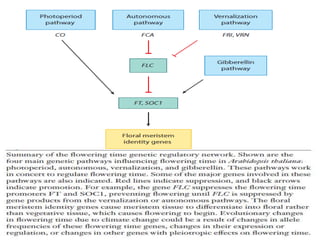
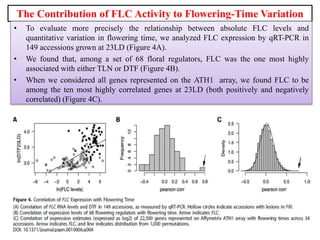
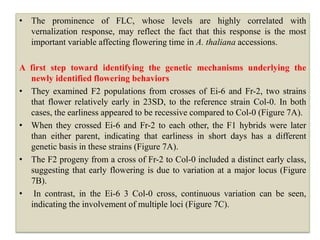
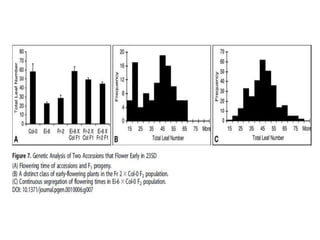
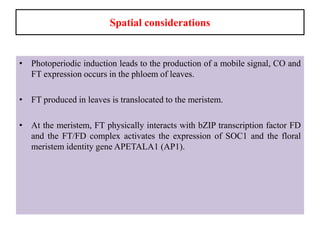
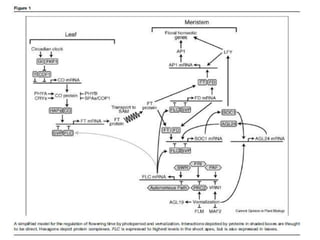
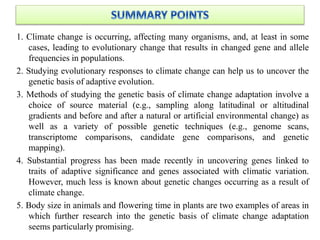
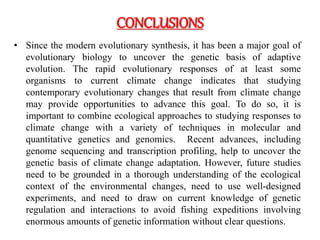
Climate change is causing rapid changes to global temperatures and precipitation patterns. This document discusses the genetic effects of climate change and methods to identify the genetic basis of adaptive evolution in response to climate change. Genome scans, transcriptome analysis, and examining candidate genes can be used to compare populations from different climatic conditions and identify genes related to climate adaptation. Studies in boreal black spruce and Arabidopsis thaliana have identified genes with single nucleotide polymorphisms associated with temperature and precipitation variables, providing candidate genes for climate change adaptation.