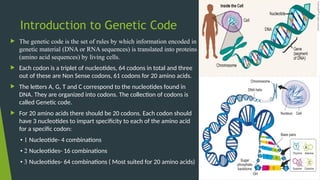
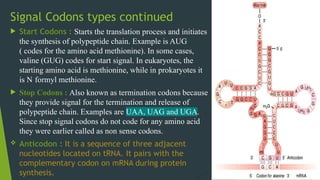
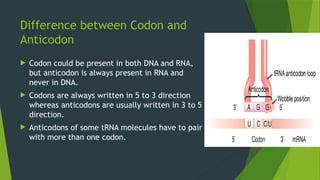
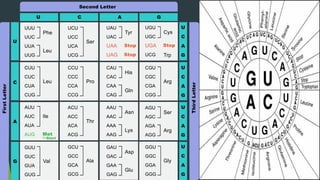
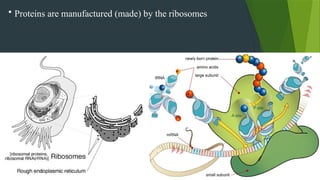
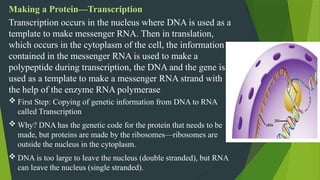
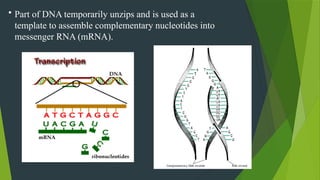
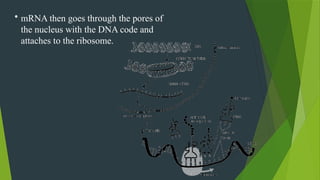
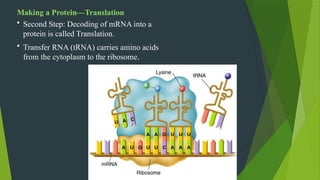
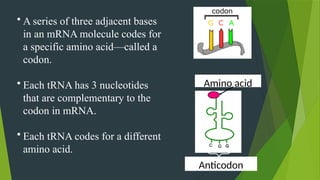
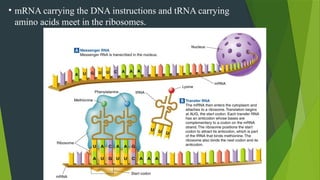
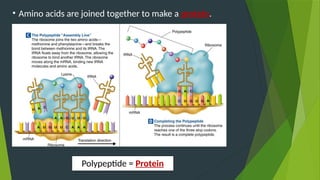
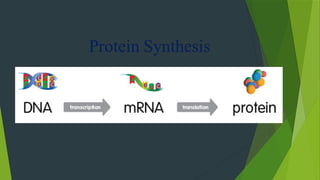
The document discusses the genetic code and protein biosynthesis, detailing the structure and function of codons, including types of genetic codes and their properties. It explains the transcription and translation processes involved in synthesizing proteins from genetic material, highlighting the roles of mRNA and tRNA. The content includes references to key studies that contribute to our understanding of the genetic code.