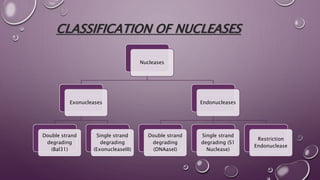
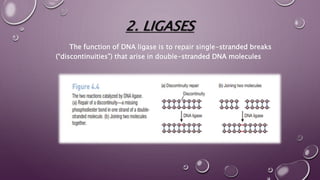
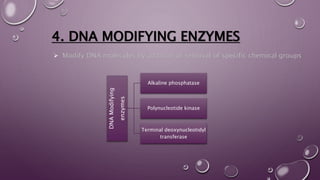
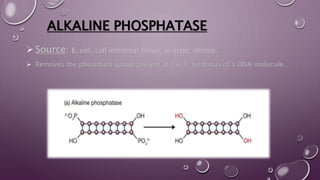
This document discusses gene cloning steps and DNA manipulation techniques. It describes how DNA can be cut, joined, copied, and modified using various purified enzymes like nucleases, ligases, polymerases, and modifying enzymes. Nucleases cut DNA, ligases join DNA fragments, polymerases make copies of DNA, and modifying enzymes add or remove chemical groups from DNA. Common nucleases discussed are restriction endonucleases, exonucleases, and endonucleases. DNA manipulation techniques allow for gene cloning, studying DNA biochemistry, and gene expression studies.