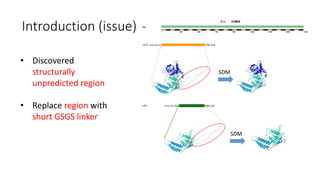
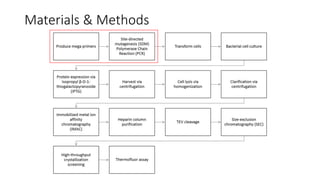
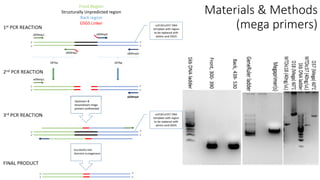
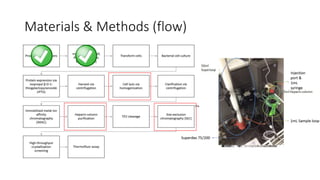
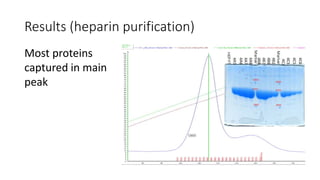
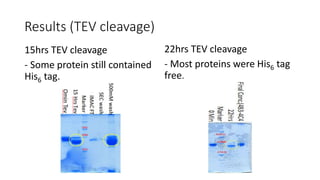
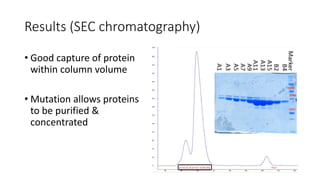
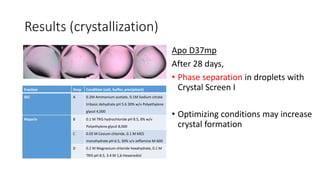
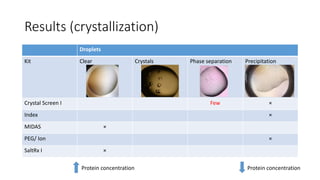
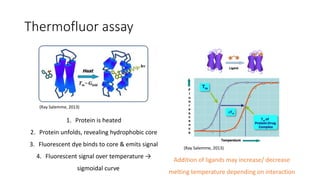
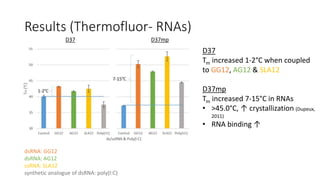
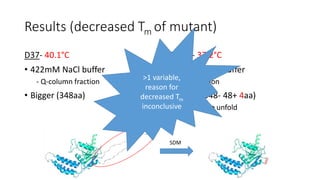
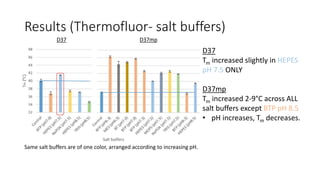
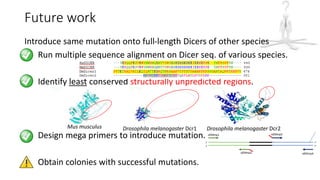
This document summarizes research on expressing, purifying, and screening for crystallization of the human Dicer helicase protein. It describes how a structurally unpredicted region in the protein construct was causing purification issues. By replacing this region with a short linker via site-directed mutagenesis, the mutant protein could now be purified and concentrated for further study. Thermofluor assays showed the mutant protein had higher melting temperatures when bound to RNA, improving its potential for crystallization. Various crystallization screening conditions were tested on the mutant protein, with some droplets showing phase separation and potential for crystal formation upon optimization. Future work proposed introducing the same mutation to Dicer proteins from other species to aid structural determination.



![References
• Salemme, R. (2013). B1. Label Free Drug Screening. [online] Available at: http://www.beta-
sheet.org/page11/page17/page18/index.html [Accessed 2 Feb. 2015].
• Salemme, R. (2013). E. Thermofluor Updates. [online] Available at: http://www.beta-
sheet.org/page11/page45/index.html [Accessed 2 Feb. 2015].
• Dupeux, F., M. Rower, G. Seroul, D. Blot and J.A Marquez. (2011) A thermal stability assay can help
to estimate the crystallization likelihood of biological samples. CrossMark, D67, 915-919
• n.a., (2015). Hampton Research. [online] Available at:
http://hamptonresearch.com/tip_detail.aspx?id=150 [Accessed 16 Jan. 2015].
• Soding, J., A. Biegert and A.N. Lupas. (2005) The HHpred interactive server for protein homology
detection and structure prediction. Nucleic Acids Res, 33, W244-248.
• Wostenberg, C., J.W. Lary, D. Sahu, R. Acevedo, K.A. Quarles, J.L. Cole and S.A. Showalter. (2012)
The Role of Human Dicer-dsRBD in Processing Small Regulatory RNAs. PLOS ONE, 7 (12), 1-12.
• Yale.edu, (2015). [online] Available at:
http://www.yale.edu/giraldezlab/miRNA_files/shapeimage_4.png [Accessed 3 Feb. 2015].](https://image.slidesharecdn.com/85a51e4a-226f-4087-8f4c-2e293a6b4b97-150303045534-conversion-gate01/85/FYP-Internal-Presentation-COMPILED-22-320.jpg)