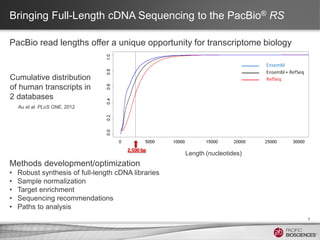
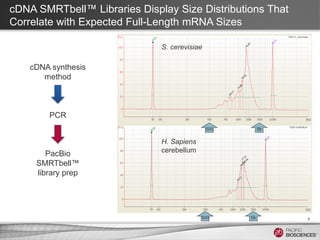
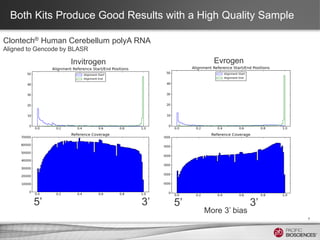
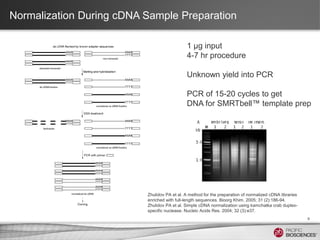
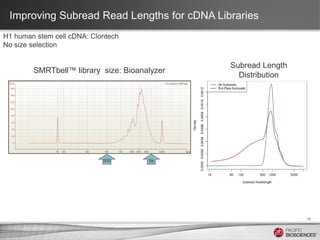
This document discusses methods for sequencing full-length cDNA using the PacBio RS system. It compares two common cDNA library preparation kits and shows they produce libraries with expected size distributions but differ in input requirements and stringency. It also demonstrates that normalization during cDNA preparation increases coverage breadth by detecting more genes. Size selection or targeted enrichment methods like SureSelect can be used to focus on specific transcript sizes or gene subsets. Analysis options include aligning reads to transcript or genome references to characterize isoforms or detect novel splice variants.