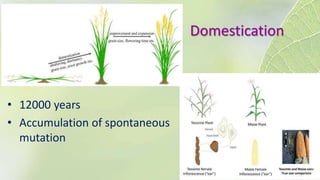
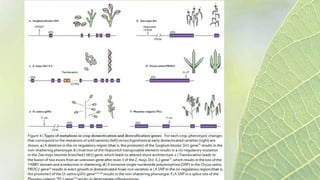
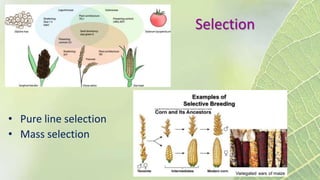
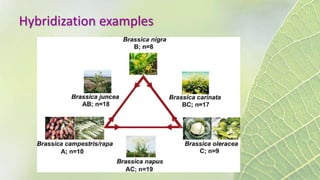
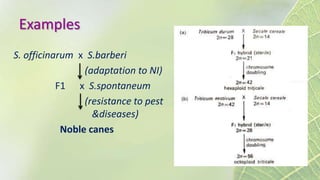
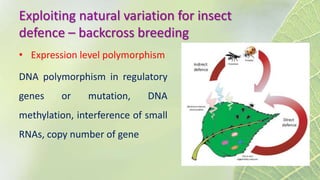
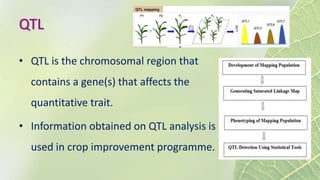
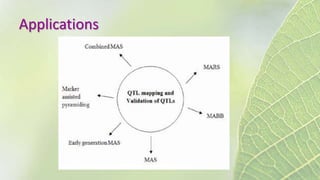
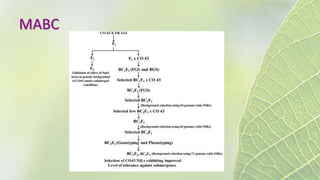
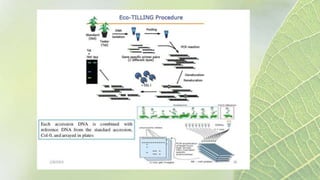
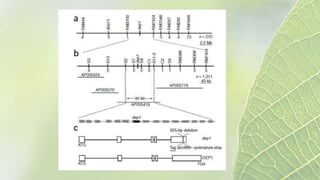
The document discusses the exploitation of natural variation in plants for improvement, focusing on genome-encoded phenotypic differences that aid adaptation and species formation. It details early domestication techniques, including selection and hybridization, emphasizing the genetic diversity found in landraces and their role as gene reservoirs. Additionally, it covers advanced methods like backcross breeding and QTL analysis that enhance crop traits such as pest resistance and grain yield.