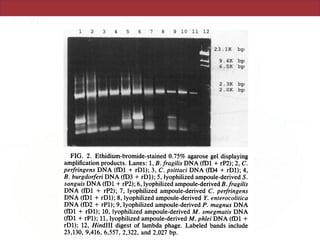
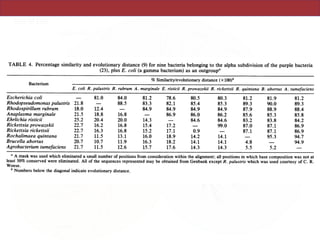
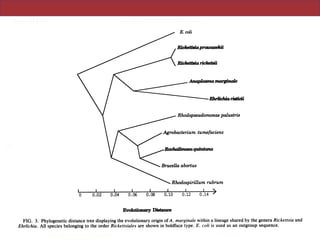
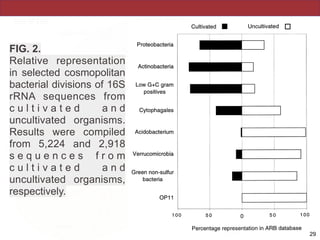
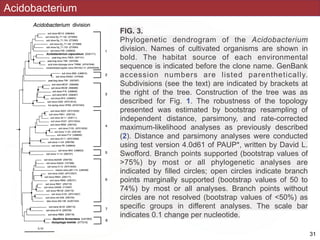
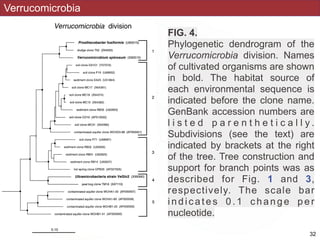
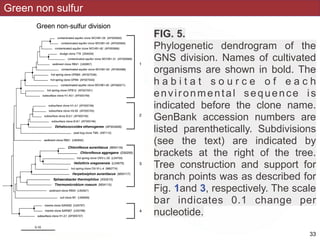
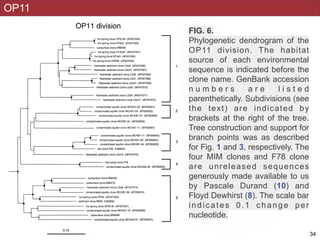
This document describes a study that used polymerase chain reaction (PCR) to amplify 16S ribosomal DNA (rDNA) from various bacterial species for phylogenetic analysis. The researchers designed universal primers that could amplify nearly full-length 16S rDNA from many bacterial genera. They demonstrated that this method allowed phylogenetic analysis of fastidious or pathogenic bacteria directly from lyophilized cultures without requiring cultivation. As an example, they amplified, cloned, sequenced and phylogenetically analyzed the 16S rDNA of Anaplasma marginale, placing it within the genera Rickettsia and Ehrlichia.