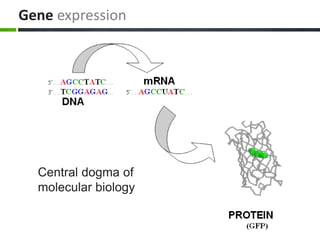
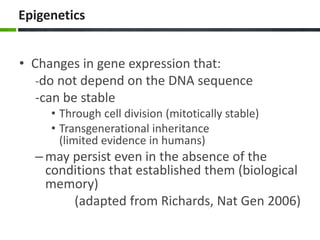
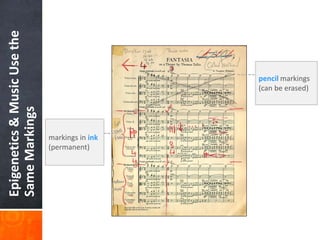
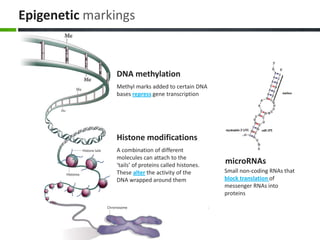
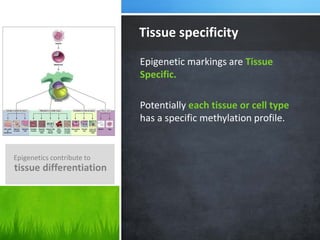
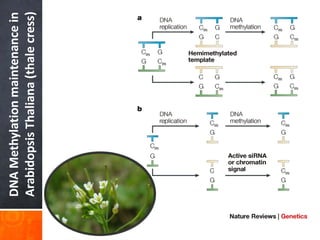
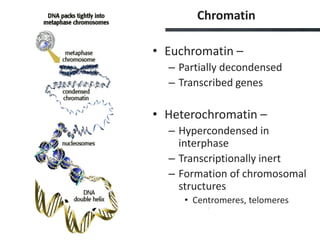
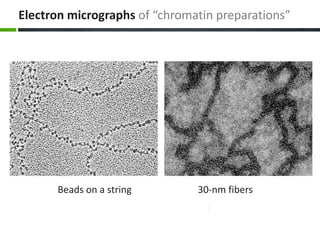
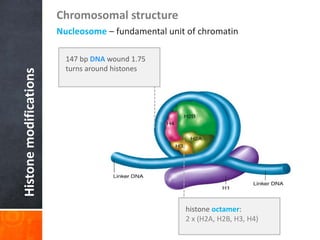
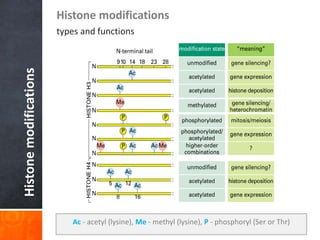
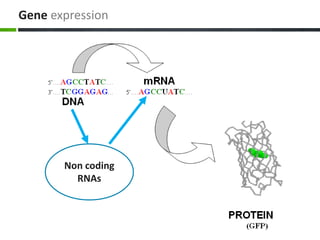
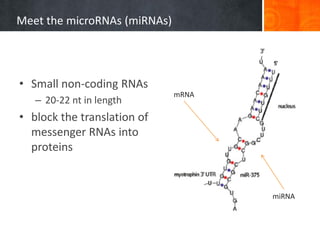
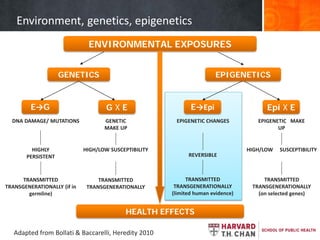
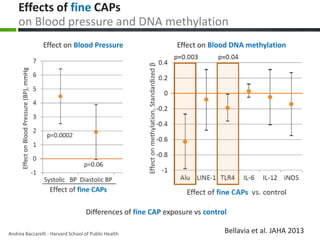
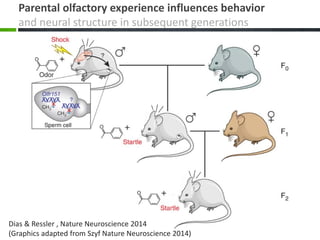
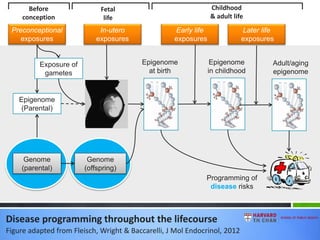
This document provides an overview of epigenetics concepts and environmental influences on epigenetics. It begins with introductory definitions of epigenetics and mechanisms like DNA methylation and histone modifications. It then discusses how the environment can influence the epigenome and provides examples like how air pollution exposure is associated with changes in DNA methylation and blood pressure. It also discusses how parental experiences like odor exposure can influence offspring behavior and neural structure through epigenetic changes in sperm. The document concludes by discussing epigenome-wide study approaches to investigate associations between exposures, epigenetic marks, and disease across the genome.
















![Examples (DNA methylation)
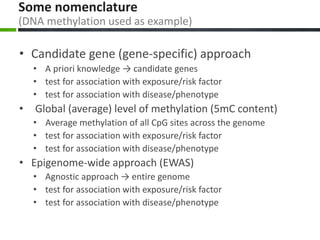
• Candidate gene approach
– Participant #1’s blood has 26% methylation in the IL6
promoter (N.B.: any other region of interest can be
targeted, e.g., CpGi shore, shelf, etc.)
• Global methylation approach
– Participant #1’s blood has 4.5% methylation (i.e., 4.5% of
all cytosines found in blood are methylated; no
information on where the methylated cytosines are
located)
• Genome-wide approach
– Methylation in Participant #1’s blood is measured at a high
number of CpG sites (e.g, if we use Illumina Infinium 450K
beadchip → we will get ≈486,000 numbers [one for each
CpG site] for Participant #1’s blood)](https://image.slidesharecdn.com/epigenetics101-baccarelli-240329083003-e549c424/85/EPIGENETICS101-BACCARELLI-EPIGENETICS101-BACCARELLI-PDF-46-320.jpg)