This paper discusses the classification of genetic mutations in cancer through machine learning, specifically using Naive Bayes and logistic regression algorithms. It outlines the challenges in manually diagnosing cancer mutations and presents a structured approach to improve classification efficiency, leveraging datasets from Kaggle. The research emphasizes the importance of accuracy and precision in predictions to facilitate personalized cancer treatment.
![Classification of Genetic Mutations Using Naive Bayes and Logistic Regression Machine
Learning Approaches
Abstract
Cancer is among the top chronic disease leading to the cause of enormous deaths in Africa. It’s
diagnosis and treatment are still difficult tasks to maneuver. Due to this fact, it has influenced
millions to lose their lives. Recently, personalized medicine has been developed to cater to the
patients who have been diagnosed this cancer by the pathologists. Despite the fact of this
development, the time taken to diagnose and develop an understanding of genetic mutations in
cancer tumors so as to make the right prescriptions for patients involves a lot of manual work and
it takes long, giving chances for this cancer tumor to develop severely. This manual system is
based more on clinical texts that must be read and understood by the pathologist so that he or she
can provide a specific therapy. This paper addresses this challenge with an approach of
constructing a machine learning model with a hope to improve the performance of classification
of genetic mutations and know the class of cancer for a proper personalized medicine therapy.
Keywords: K-Nearest Neighbor; Naive Bayes; Classifier; Cancer; Personalized Medicine.
I. INTRODUCTION
In Africa, Uganda has registered over 22,000 Cancer death cases by 2018[1], and these figures are
doubling every year. Cancer develops from the long-lived cells in a human body that multiply out
of control. A human body is composed of over 80 trillion cells, of which any of these cells can
cause cancer when it behaves abnormally. There a numerous kind of genes found inside these cells
that make up a human being. Cancer is a very complex chronic disease to treat due to its nature
and this makes its treatment gradually very slow. Personalized medicine has mainly involved the
orderly use of any other information concerning a patient to develop that patient’s pre-emptive and
healing care[2, 3]. Information concerning individuals’ or patients’ genetic profile that can be used
to provide the right specific treatment for the patient. However, for the effectiveness of
personalized medicine, it requires deeply analyzing the proper cause of cancer and its stream of
how it can be treatment. Traditionally, the patient biomedical data is analyzed manually. It
involves high dimension data greater than 3 which makes it extremely difficult and often
impossible to much and come up with the correct personalized medicine for the patient[4].
Additionally either genetic mutations or neural mutations has an upper-hand to the development
or cause of cancer, or facilitating tumor growth[3]. This has no proper evidence and requires a lot
of research to analyze the major cause. For a newly created cancer tumor, it can bear hundreds or
thousands of genetic mutations. Apparently, the process of distinguishing these genetic mutations
is being done manually by clinical pathologists, this process is a slow very time-consuming task
where a clinical pathologist has to manually review thousands of research notations and papers
available for each genetic mutation and classify every single genetic mutation based on evidence
so as to understand the type of cancer[3]. This problem has been solved before using different
classification algorithms[3]. But for our case, we shall use the Naive Bayes and Logistic
Regression machine learning algorithms to provide solution and also bring significant
improvement in sequencing the provided datasets properly[5].](https://image.slidesharecdn.com/definecancertreatmentusingknnandnaivebayesalgorithms-200114073723/75/Define-cancer-treatment-using-knn-and-naive-bayes-algorithms-1-2048.jpg)
![This paper is organized as follows. Section II describes approaches used with experimental dataset.
Results of these approaches are also presented in Section III. Section IV analyzes the obtained
results briefly. Then lastly, Section V marks a conclusion of this paper and further works.
II. Research methods
Machine learning is a subset of artificial intelligence, refers to the ability of IT systems to
independently develop solutions to problems by recognizing patterns in databases basing of
existing algorithms and data sets and to develop adequate solution concepts[6]. Using machine
learning, the software can independently generate solutions once data has been fed into the systems
and can perform the various tasks by Machine Learning like; finding, extracting and summarizing
relevant data, making predictions based on the analysis data, calculating probabilities for specific
results, adapting to certain developments autonomously, optimizing processes based on recognized
patterns.
In this research the major task is to classify genetic mutations to enable the right personalized
medicine for cancer treatment. The datasets are obtained from a publicly available source” Kaggle”
and it’s called “Personalized medicine: Redefining Cancer treatment”[7]. Initially, We analyzed
and understood the two datasets (training_variants and training_text datasets), secondly,
understood the same problem from the machine learning perspective, identified the kind of data
present in the class column of the training_variants dataset (discrete data) and so it’s a
classification problem because of the multi-discrete output. The nature of the datasets was a text-
based, therefore, converting the datasets was a must. We used Naive Bayes and Multinomial
Logistic Regression for classification. Lastly, evaluation of two classification methods was done
basing on the multi class log loss and accuracy of the models.
III. Approach
The same problem was addressed before using Support Vector Machine and XGBoost
Classification algorithms[3]. Our approach to this problem basically involves two major steps;
first; data cleaning, transformation and extraction of relevant features in the training set for training
and Secondly, implementation of classification methods (Naive Bayes and Logistic regression) to
develop the model. To solve this medical cancer problem, three variables where required; gene,
variation and the clinical_text so that we can predict the class to which the specific type of cancer
belongs to. But unfortunately, the clinical_text evidence that came with related medical literatures
is in text format, it couldn’t be fed into the machine learning algorithm, therefore, we convert and
extract it in a format that ML algorithm can deal with it as shown in fig.1 below;](https://image.slidesharecdn.com/definecancertreatmentusingknnandnaivebayesalgorithms-200114073723/75/Define-cancer-treatment-using-knn-and-naive-bayes-algorithms-2-2048.jpg)
![Fig. 1. Illustration of the approach to solve the medical problem
III. Experiments
A. Datasets
Both datasets where obtained from Kaggle competition and Memorial Sloan Kettering Cancer
Center (MSKC)[7]. The training_variants dataset has four columns (ID, Gene, Variation and
Class), and the training_text dataset has two attributes (ID and the clinic _text). To solve this
problem three variables; Gene, Variation and clinical_text variables are required to predict
mutation classes. Given any new gene, variation associated with it and the research paper
associated with it, you should be able to predict the class it belongs to. In the training_ variants
dataset we have 3321 records with 4 features and in the training_variants we have 3321 records
with 2 features. In the training_variants nine classes of mutations are given.
This is a medical rated problem errors are very costly; therefore, accuracy is highly important. The
results of each class are expected to be in terms of probability in order to build more evidence and
to be more confident when awarding results to the patient or to have a grounded reasoning why
the Machine learning algorithm is predicting a class. Since both datasets have a common column
called the ID, from it point the two datasets are merged into one common dataset after data
preprocessing. After that we formed one common dataset having 5 features (ID, Gene, Variation,
Class and clinical_text). But there was some missing data which would impact my analysis, so we
performed some imputation (replaced missing data with substituted ‘gene’ value), since they are
small in number.
B. SPLINTING DATA IN TRAIN, VALIDATION AND TEST SETS
After data enrichment and data wrangling, i splinted our newly formed into train, validation and
test sets. It’s extremely important to split our data because when you build your model using the
whole data, then there is no way your model will be validated. We built and verified our model
using the training and cross-validation sets. Cross-validation was basically used for
hyperparameter tuning or optimization and finally validated the model output using the test set.
C. DATA DISTRIBUTION IN TRAIN, VALIDATION AND TEST SETS
We look into the distribution of different genetic mutation classes in train, cross-validation and
test sets in figures 1, 2 and 3 respectively.](https://image.slidesharecdn.com/definecancertreatmentusingknnandnaivebayesalgorithms-200114073723/75/Define-cancer-treatment-using-knn-and-naive-bayes-algorithms-3-2048.jpg)
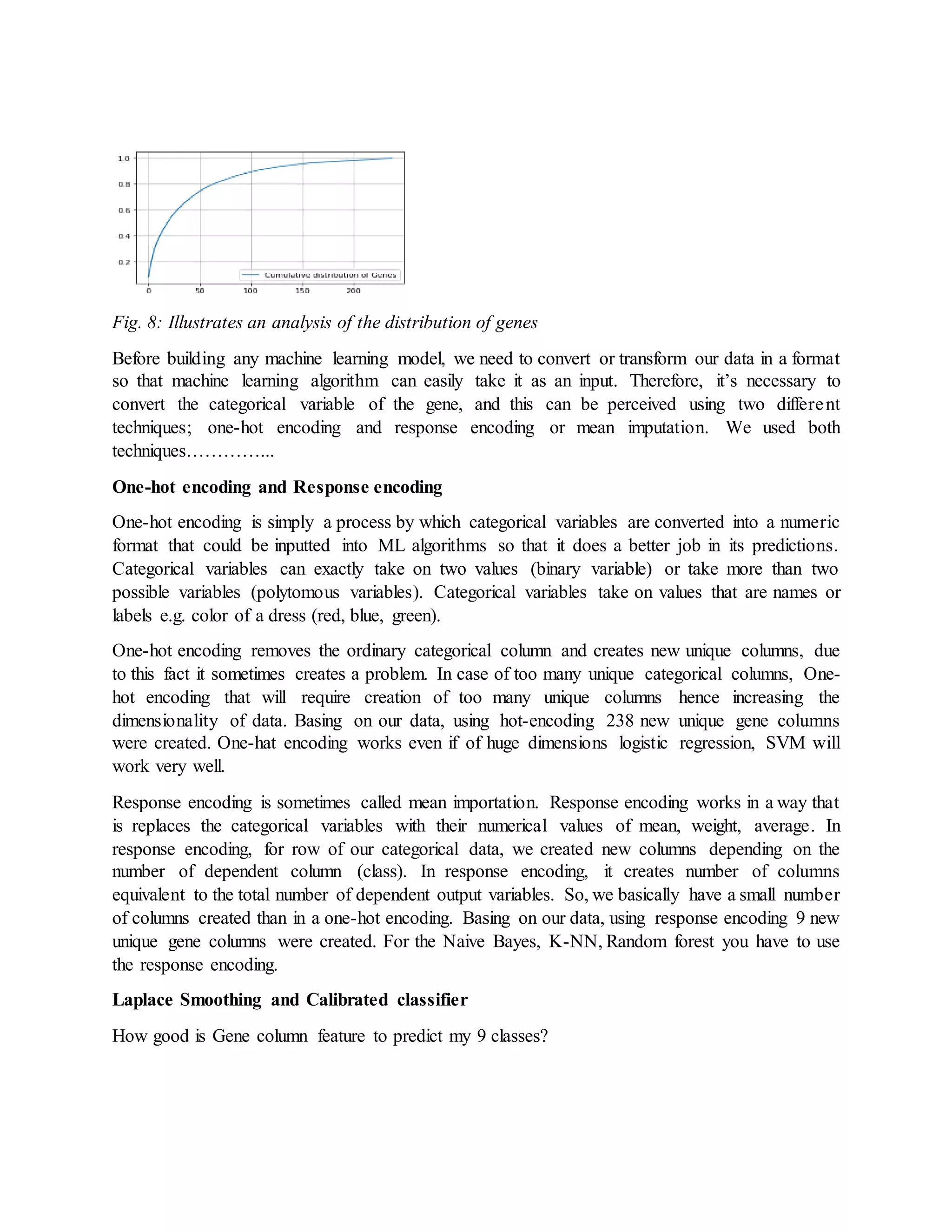
![EVALUATION
Various ways can be used to evaluate a ML model, accuracy, area under a curve among many
other evaluation matrices. But i evaluated our model using the multi class log loss or sometimes
called the cross-entropy loss and a Confusion matrix.
LOG LOSS, MULTI CLASS LOG LOSS OR CROSS ENTROPY.
Log loss stands for logarithmic loss, its values range from 0 to ∞, it’s a loss function for
classification that quantifies the price paid for inaccuracy of predictions in classification
problems[8]. Log loss is used for binary classification algorithms (limited to only two possible
outcomes). Additionally, for a perfect model its log loss should be zero. But in real world there is
no perfect model. If more of your prediction is better, the log loss value is going to be low. And if
your prediction is ambiguous or unclear the log loss will penalize you. Therefore, the main idea
behind log loss is to keep the value low.
Log loss penalizes false classifications by taking into account the probability of classification.
Mathematical formula of log loss;
𝒍𝒐𝒈 𝒍𝒐𝒔𝒔 = −
1
𝑛
∑[𝑦𝑖 log 𝑝𝑖 + (1 − 𝑦𝑖)log(1 − 𝑝𝑖)]
𝑛
𝑖=1
Where n represents the number of samples or entities, 𝒑𝒊 represents the possible probability, 𝒚 𝒊
represents the Boolean original outcome in the i-th instance.
Multiclass log loss or cross entropy values, range from 0 to ∞, it’s a loss function for classification
that quantifies the price paid for inaccuracy of predictions in classification problems, it penalizes
false classifications used for multiclass classification.
Mathematical formula of cross entropy;
𝑪𝒓𝒐𝒔𝒔 𝒆𝒏𝒕𝒓𝒐𝒑𝒚 𝒍𝒐𝒔𝒔 = −
1
𝑛
∑ ∑ 𝑦𝑖 𝑗
𝑐
𝑗=1
𝑛
𝑖=1
log(𝑝𝑖 𝑗 )
Where c represents the number of classes, n represents the number of samples or entities, 𝒑𝒊
represents the possible probability, 𝒚 𝒊 represents the original outcome in the i-th instance. 𝒑𝒊 𝒋 is
the model’s probability of assigning label j to instance i.
BUILDING A WORST-CASE MODEL.
Accuracy ranges from 0 to 1, having an accuracy closer to 1 is great e.g. if it’s 0.95, the accuracy
will be 95% accuracy. In log loss a value can go from 0 to ∞. If the value comes out to be 0 then
that marks it to be a best model. Assuming we get a log loss value greater than 1, how can we
evaluate our model whether it’s a good or bad model. There a various method but for our case we
used a random model so called worst model. This model was created by completing some 9 random
numbers on our dataset because we have 9 classes, the sum should be equal to 1 because their sum
of probability is equal to 1. We generated the log loss for all sets (train, validation and test sets).](https://image.slidesharecdn.com/definecancertreatmentusingknnandnaivebayesalgorithms-200114073723/75/Define-cancer-treatment-using-knn-and-naive-bayes-algorithms-5-2048.jpg)





![So, we need to bring all the three independent columns together to work and build the model (gene,
variation and the text). Using both one-hot encoding and response encoding we obtained the
following shapes for all three kinds of datasets as illustrated in the figures….. below;
we observe that we got much greater number of columns when using the one-hot encoding with
values of 55659 columns since we have 18553 number of columns in each set of the three sets.
We have 27 columns when using the response encoding feature because each set has 9 unique
classes.
Building the machine learning models
I. Naive Bayes Classification Model.
Since we have a lot of text data, we decided to start building our model with Naive Bayes
classifier. Naive Bayes is a probabilistic classifier based on applying the Bayes theorem[9], it’s
also known as simple Bayes and independence Bayes. We built our model using all the three
independent columns (gene, variation and text columns).
Bayes theorem:
𝑃( 𝐴𝐵) =
𝑃( 𝐵𝐴) ∗ 𝑃(𝐴)
𝑃(𝐵)
Using Bayes theorem, we can find the probability of A happening, given that B has occurred.
Accordingly, B is the evidence and A is the hypothesis. The assumption made are, the predictors
or features are independent, (presence of one particular feature does not affect the other). Naive
Bayes classifier converge quicker than any other discriminative models like logistic regression, so
you it requires a small amount of training data[10].
We used the alpha and the multinomial Naive Bayes classifier for multiclass classification. We fit
the model using the one-hot encoding. We then get the log loss of alpha values with a minimum
value of 1.27 at alpha 0.1 as illustrated in the figure….. shown below;](https://image.slidesharecdn.com/definecancertreatmentusingknnandnaivebayesalgorithms-200114073723/75/Define-cancer-treatment-using-knn-and-naive-bayes-algorithms-12-2048.jpg)


![(9.2%), this is a big difference, so there is no confusion hence the model is working very well. And
this answers the above question.
The accuracy of our model;
𝐴𝑐𝑐𝑢𝑟𝑎𝑐𝑦 = 1 − 𝑁𝑢𝑚𝑏𝑒𝑟 𝑜𝑓 𝑚𝑖𝑠𝑐𝑙𝑎𝑠𝑠𝑖𝑓𝑖𝑒𝑑 𝑝𝑜𝑖𝑛𝑡𝑠
= 1 − 0.3890977443609023
= 0.610902255
𝐴𝑐𝑐𝑢𝑟𝑎𝑐𝑦 = 61.1%
This was quite a good accuracy according to the problem we were solving. Therefore, the output
results for our model were quite good with such an impressive percentage of accuracy.
II. Logistic regression
Logistic regression describes data, and explains the relationship between one dependent binary
variable and one or more independent categorical variables sometimes called the target[11]. There
are basically three types of logistic regression; binary, multinomial and ordinal LR. Since the
problem we are solving it a multinomial problem we shall concentrate on multinomial LR.
We are going to do over sampling because in most classes we had much data and in some few
classes we had less data, due to this factor it can affect the performance of our model. We shall try
to balance the classes. Here we try to demonstrate our model with balanced classes.
Balanced classes
Using the SGD Classifier, we balanced our classes. We got the minimum log loss value of 1.19 at
alpha point 0.001 also demonstrated in the figure…… shown below. interpretability
We tested our log loss minimum value on the testing data using the alpha value. Our outputted log
loss value was 1.19 and the misclassified points was 0.36. From these results we can tell the
accuracy of our model. The performance of our classification model on the testing dataset was
described by a confusion matrix shown in figure…… below;](https://image.slidesharecdn.com/definecancertreatmentusingknnandnaivebayesalgorithms-200114073723/75/Define-cancer-treatment-using-knn-and-naive-bayes-algorithms-15-2048.jpg)


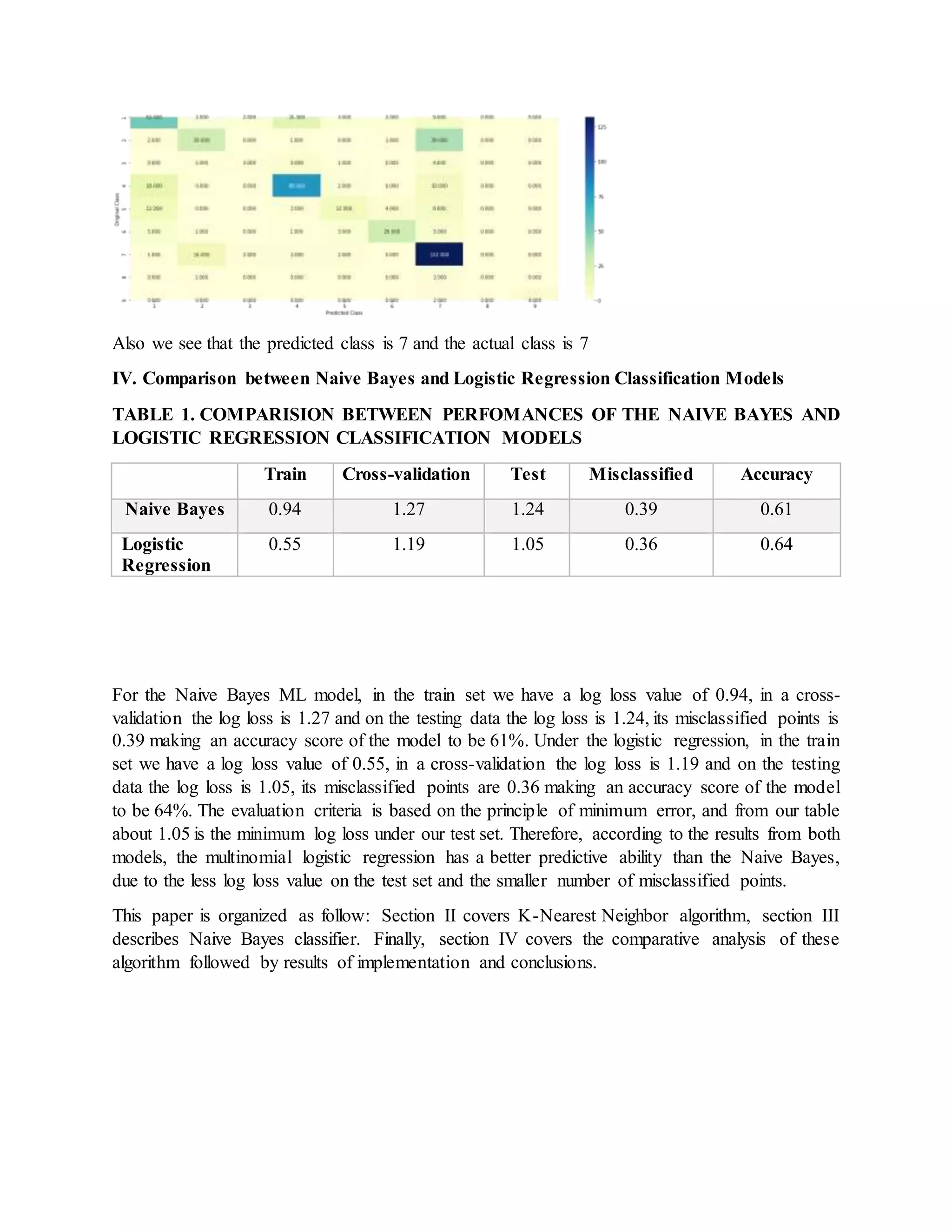
![Discussion
The preliminary results obtained from our study really verified the possibility of implanting
machine learning algorithms (Naive Bayes and Multinomial Logistic Regression) into
classification of Genetic Mutations, which is text-based and probabilistic classification problem.
This reduce deadly manual work and accelerate the progression in analyzing cancer roots. In this
section, we will further interpret this project based on the analysis of our results.
Firstly, Analyzed the shapes of our datasets, understood the independent and dependent variables
and the kind of data present in target of dependent column. It was discrete data therefore, it was a
classification problem because of the multiple discrete outputs (classes 1 to 9).
Secondly, since we had medical text data, so we preprocessed it into a good format so that ML
algorithm can understand and input it. We merged our two data sets into one dataset, then did some
imputation since we had some missing values, we then splinted our data into three sets (Train 60%,
Cross-validation 20% and Test 20%).
Thirdly, we performed random sampling since our classes (1-9) had imbalanced data so as to
improve the performance of our model.
Fourthly, we evaluated each column independently to make sure that its relevant for our target
variable (class column). Some columns had categorical data so, we converted this categorical data
using one-hot encoding technique. Finally, we observed that all the independent columns were
relevant.
Fifthly, we performed over sampling to reduce bias due to the highly twisted distributed data. And
then finally, regarding the performances of the two classification models Logistic regression
performed better than the Naive Bayes ML algorithm. Since it had fewer misclassified datapoints
and a high degree of accuracy than the Naive Bayes classifier.
Conclusion
From survey and analysis on comparison among data mining classification algorithms (Naive
Bayes and Multinomial logistic regression), it shows that logistic regression algorithm is more
accurate and has less error rate and it’s an easier algorithm as compared to Naive Bayes. When
modeling Logistic regression, we used balanced data and so we may need to also use imbalanced
data and try to compare the results obtained.
References.
[1] F. Okongo, D. M. Ogwang,B. Liu, and D. MaxwellParkin, "Cancer incidence in Northern Uganda
(2013–2016)," International journal of cancer, vol. 144, no. 12, pp. 2985-2991, 2019.
[2] M. Verma, "Personalized medicine and cancer," Journal of personalized medicine, vol. 2, no. 1,
pp. 1-14, 2012.
[3] G. Li and B. Yao, "Classification of Genetic Mutations for Cancer Treatment with Machine
Learning Approaches," InternationalJournalof Design,Analysisand ToolsforIntegratedCircuits
and Systems, vol. 7, no. 1, pp. 63-67, 2018.](https://image.slidesharecdn.com/definecancertreatmentusingknnandnaivebayesalgorithms-200114073723/75/Define-cancer-treatment-using-knn-and-naive-bayes-algorithms-19-2048.jpg)
![[4] A. Holzinger, "Trends in interactive knowledge discovery for personalized medicine: cognitive
science meets machine learning," The IEEE intelligent informatics bulletin, vol. 15, no. 1, pp. 6-
14, 2014.
[5] M. W. Libbrecht and W. S. Noble, "Machine learning applications in genetics and genomics,"
Nature Reviews Genetics, vol. 16, no. 6, pp. 321-332, 2015.
[6] A. E. Maxwell, T. A. Warner, and F. Fang, "Implementation of machine-learning classification in
remote sensing: An applied review," International Journal of Remote Sensing, vol. 39, no. 9, pp.
2784-2817, 2018.
[7] Kaggle. "Personalized Medicine: Redefining Cancer Treatment." Kaggle.
https://www.kaggle.com/c/msk-redefining-cancer-treatment (accessed.
[8] V. Vovk, "The fundamental nature of the log loss function," in Fields of Logic and Computation
II: Springer, 2015, pp. 307-318.
[9] H. Padmanaban, "Comparative analysis of Naive Bayes and tree augmented naïve Bayes models,"
2014.
[10] Z. K.Senturk and R.Kara,"Breast cancerdiagnosis via data mining: performance analysis of seven
different algorithms," Computer Science & Engineering, vol. 4, no. 1, p. 35, 2014.
[11] R. E. Wright, "Logistic regression," 1995.](https://image.slidesharecdn.com/definecancertreatmentusingknnandnaivebayesalgorithms-200114073723/75/Define-cancer-treatment-using-knn-and-naive-bayes-algorithms-20-2048.jpg)