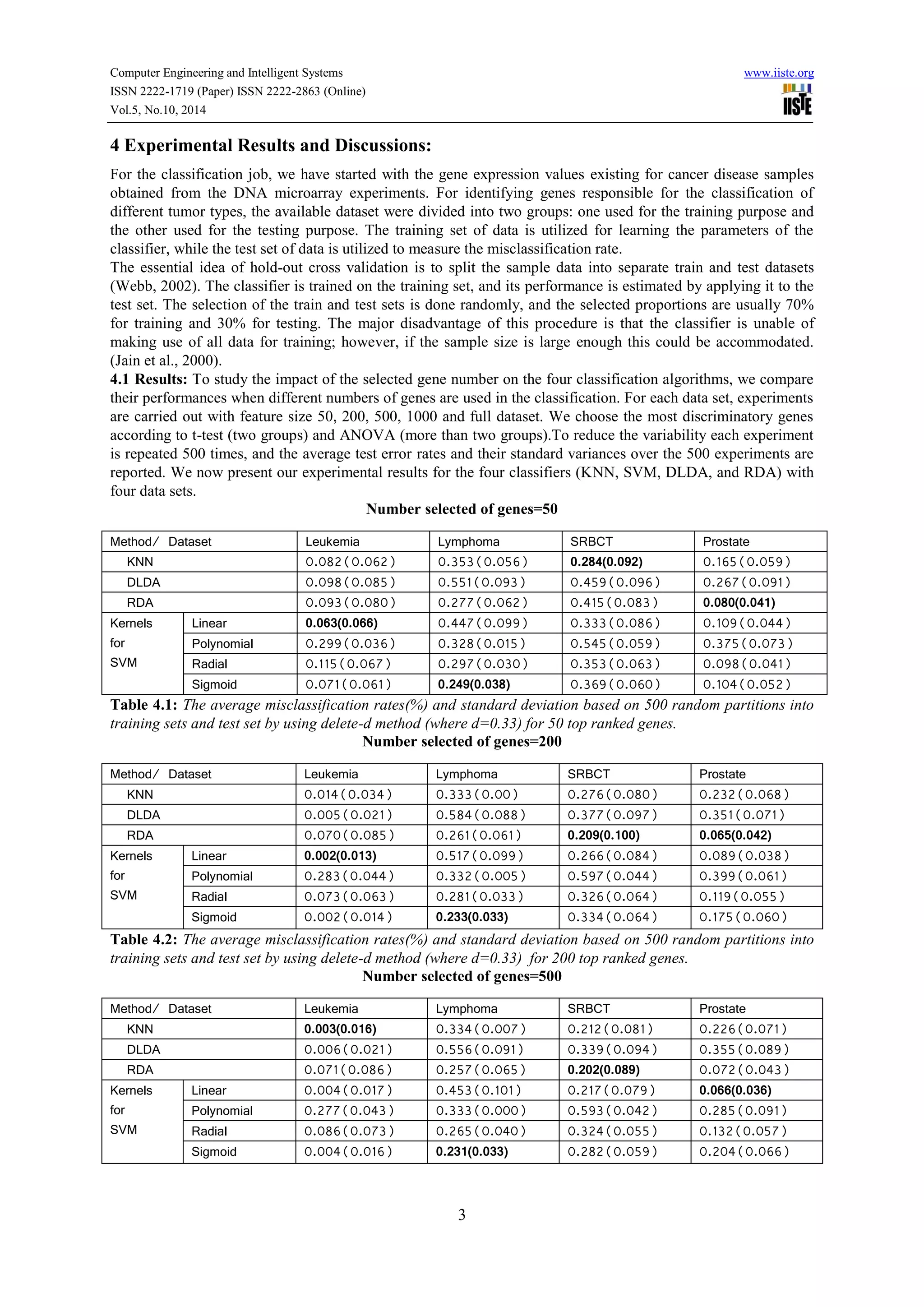
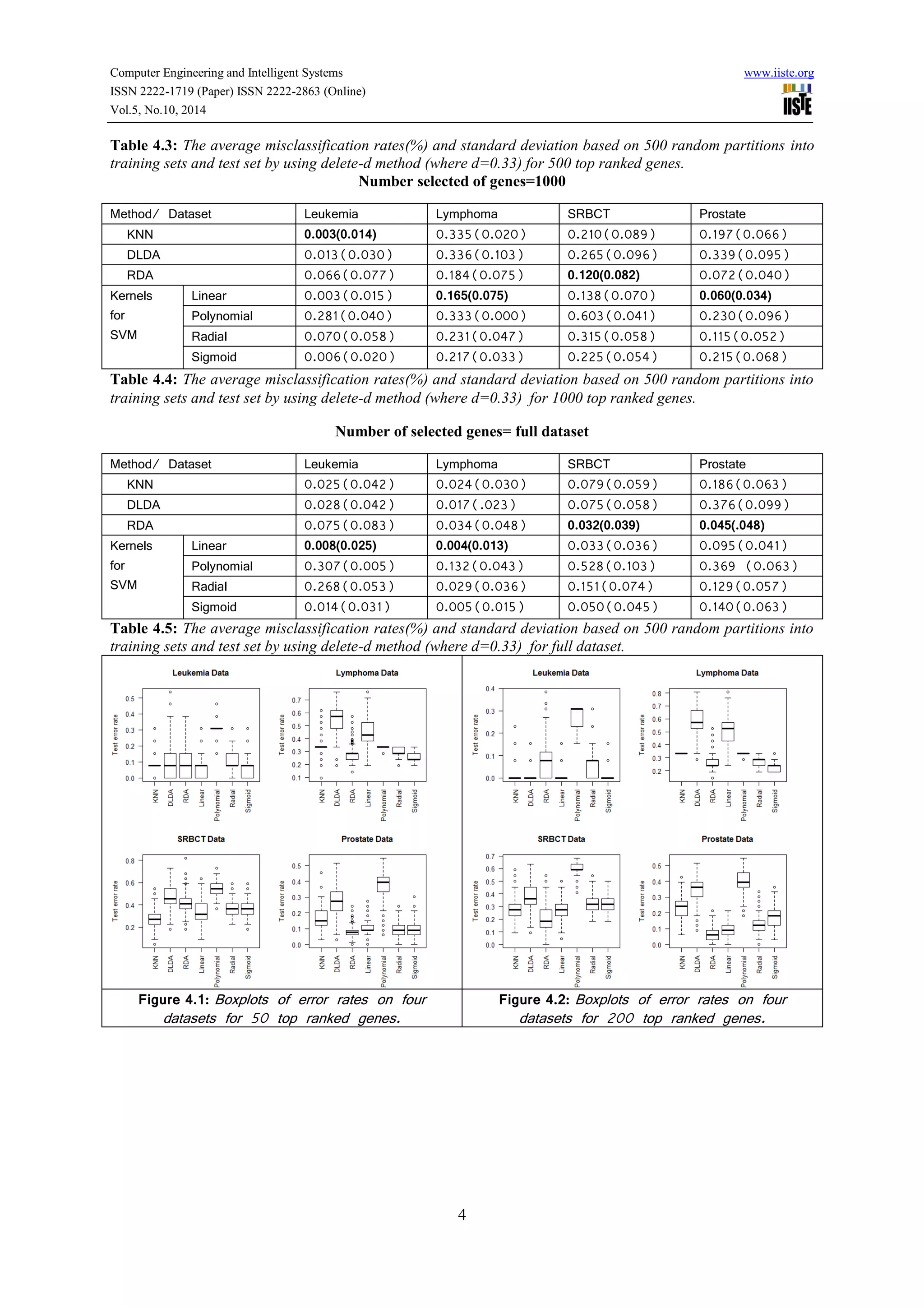
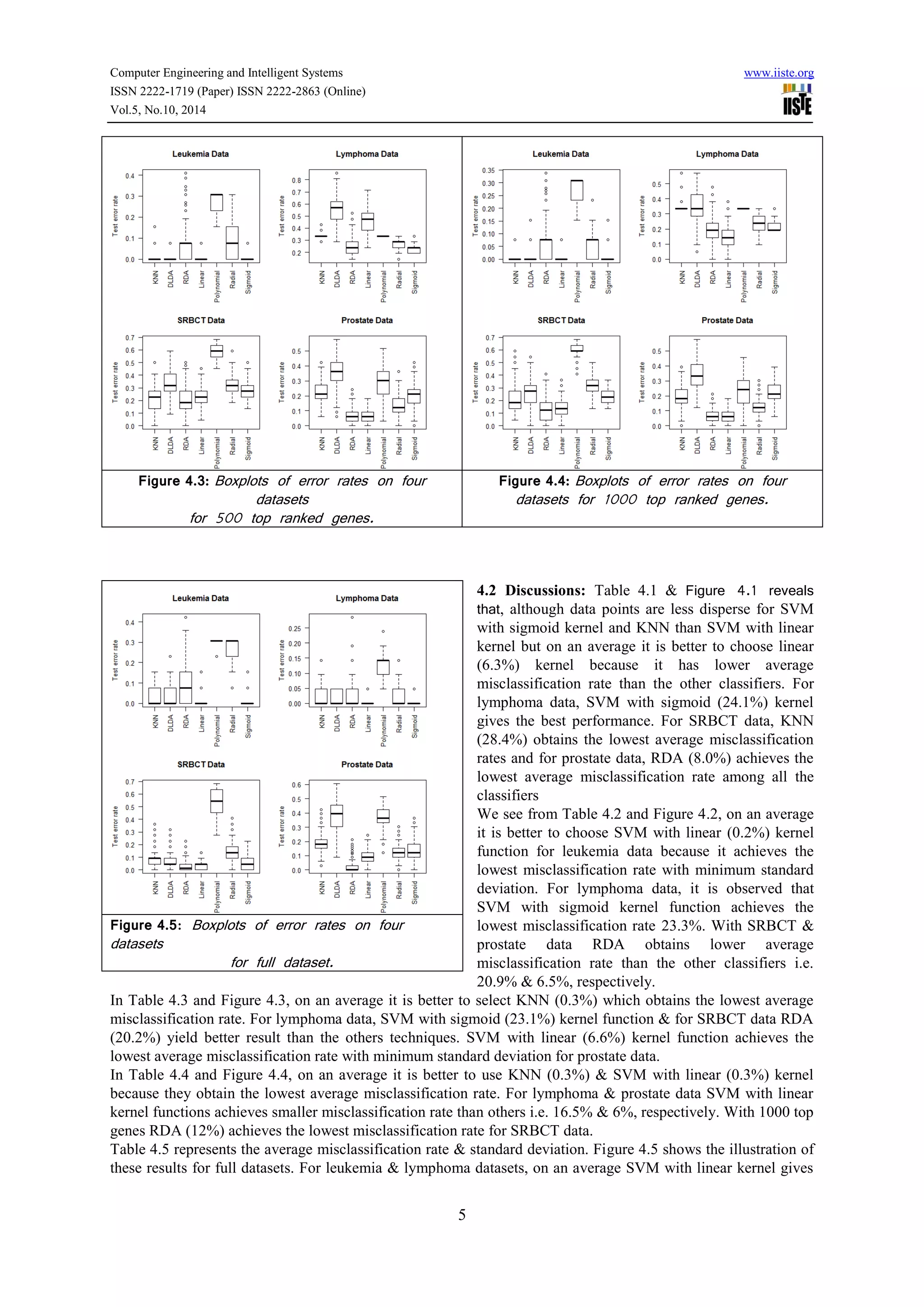
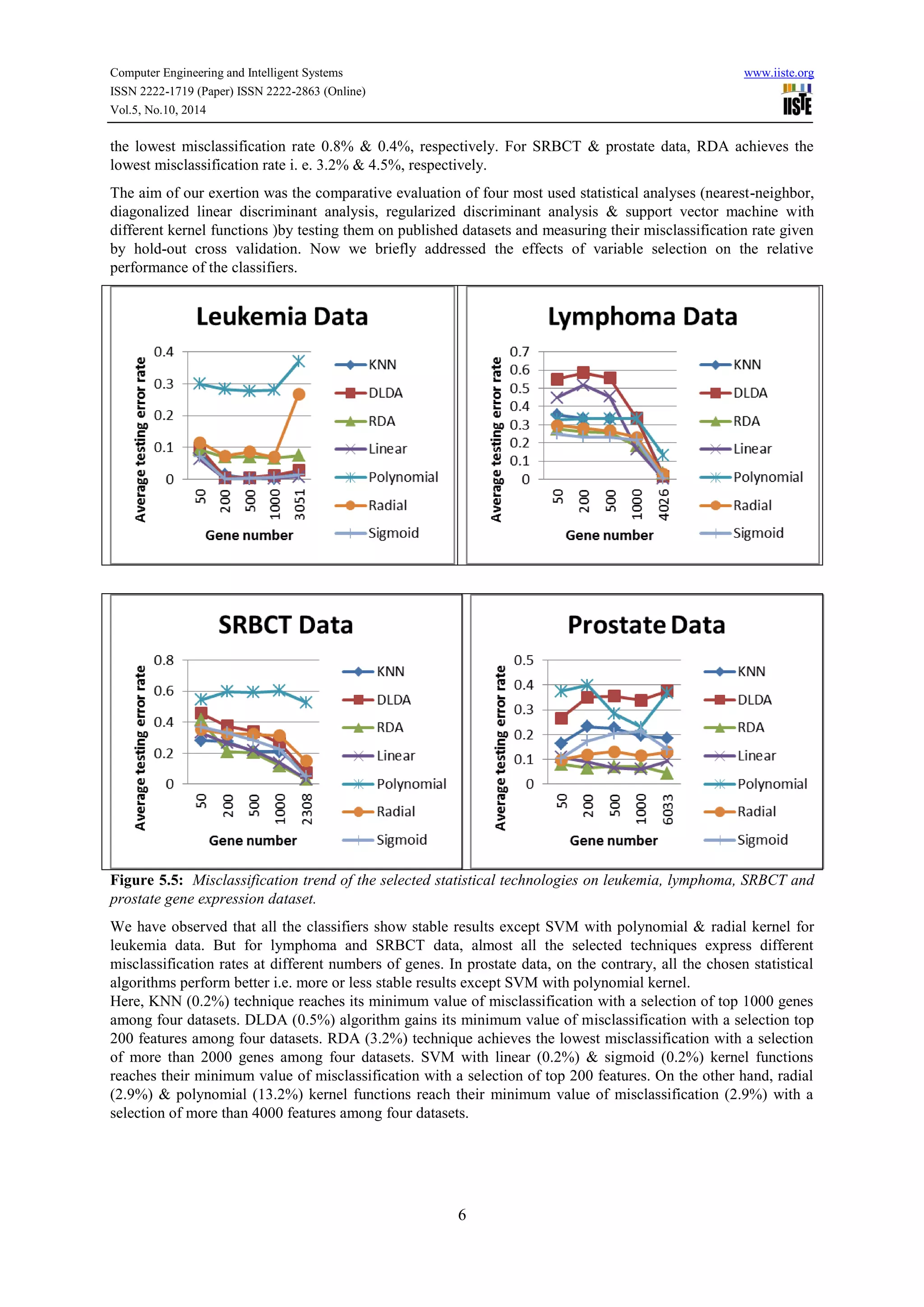
This document compares the predictive accuracy of various machine learning and statistical classification algorithms on four gene expression datasets. It finds that KNN, RDA, and SVM with a linear kernel generally have lower misclassification rates than DLDA when classifying tumors using different numbers of selected genes (50, 200, 500). Classification performance is evaluated using hold-out cross-validation, and the algorithms are tested on leukemia, lymphoma, SRBCT, and prostate cancer gene expression datasets containing between 38 and 102 samples each.