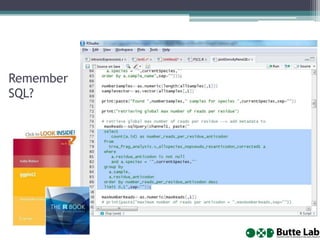
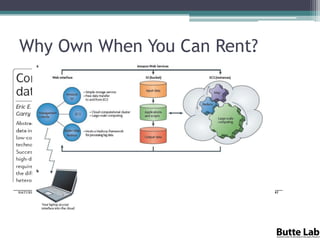
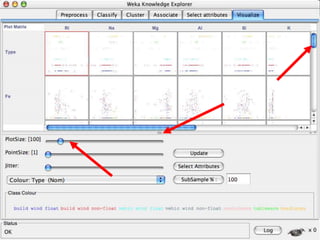
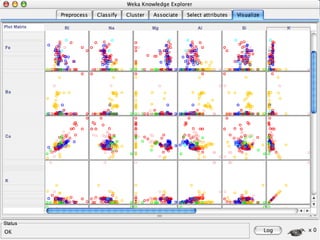
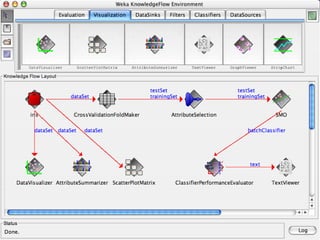
The document discusses the importance of programming skills for bioresearchers. It argues that just as pipetting skills are essential, programming skills are now equally important. It recommends moving away from solely using Excel for data storage and analysis and instead using relational databases and programming. The document outlines free and cheap software, algorithms, and cloud computing resources available to help researchers learn programming. It emphasizes that programming is necessary to address both small and large problems as off-the-shelf software will fail for real science applications.