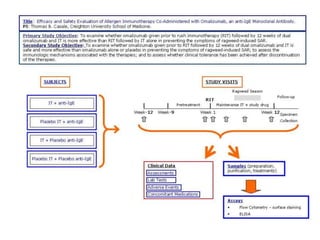
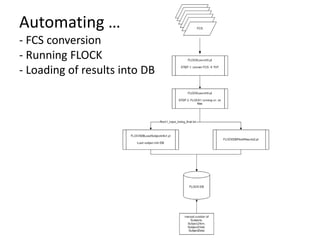
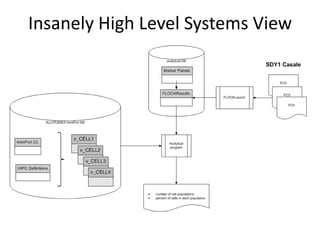
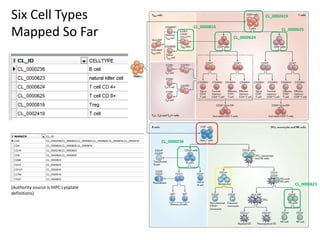
1. The document discusses automatically assigning ontological IDs to cell populations identified in studies using FLOCKMapper.
2. It describes the system components used, including a study dataset, FLOCK for analysis, the Cell Ontology and ImmPort ontologies for IDs, and HIPC definitions converted to a computable form.
3. It explains the mapping process involves identifying an ontological class that maps to a given HIPC class definition, addressing mismatches, and codifying the mappings as views and functions.