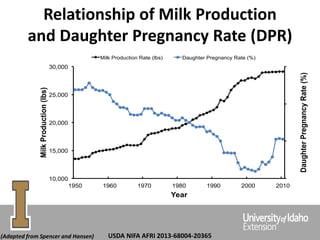
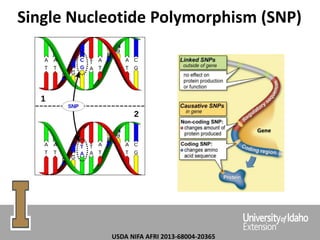
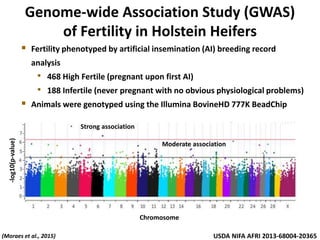
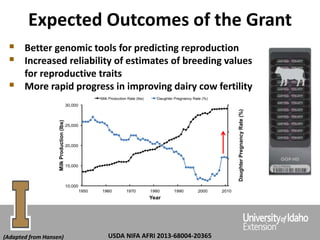
The document discusses improving dairy cattle fertility through translational genomics by identifying and utilizing genetic markers associated with fertility traits. It outlines a research approach focusing on the development of novel SNP markers and their effects on daughter pregnancy rates and embryo development, alongside efforts to disseminate this information to producers. Expected outcomes include enhanced genomic tools for predicting reproduction and increased reliability in breeding values for reproductive traits.