1) The document summarizes research on analyzing the genetic code found in cancers. It discusses how the genetic code is being decoded through advances in sequencing technology that have allowed researchers to read the 3.2 billion letters in the human genome.
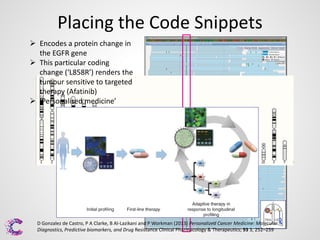
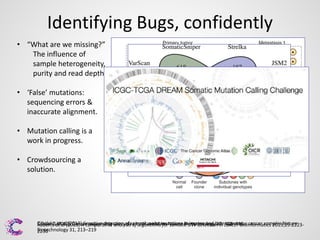
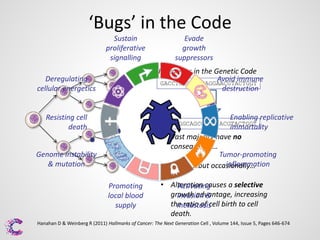
2) Mutations or "bugs" in the genetic code can cause cancers by disrupting genes involved in processes like cell growth. New sequencing methods are helping researchers precisely locate mutations and determine which may drive tumor growth.
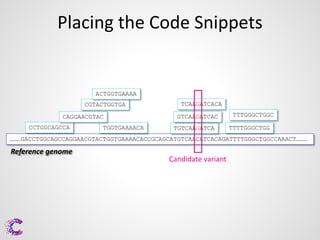
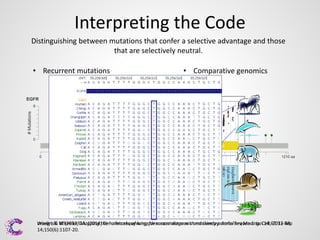
3) Interpreting the sequencing data is an ongoing challenge but identifying recurrent mutations and comparing mutations across samples and genomes can help distinguish driver mutations from harmless changes.

![Reading the Code: 1
$100,000,000
1400
1200
$10,000,000
1000
$1,000,000
800
$100,000
Tb
600
$10,000
400
$1,000
200
Cost/Genome
Short Read Archive
$100
0
2001
2002
2003
Draft Human
Genome Project
2004
2005
2006
2007
‘Massively Parralel’
sequencing
2008
2009
2010
1st Tumour
2011
2012
2013
2014
>12, 000 tumours
Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) [27/01/2014]
Wang and Wheeler (2014) Genomic Sequencing for Cancer Diagnosis and Therapy Annu. Rev. Med. 65: 33-48](https://image.slidesharecdn.com/crackingcancerscodefeb2014-140214095943-phpapp01/85/Cracking-cancers-code-feb-2014-4-320.jpg)