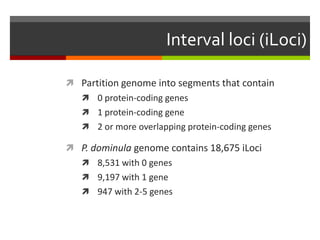
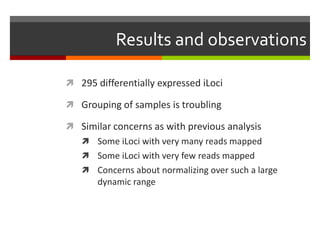
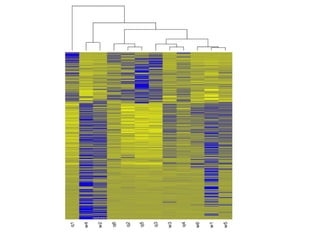
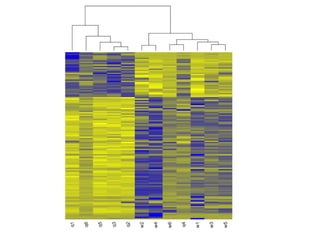
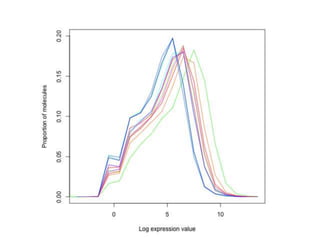
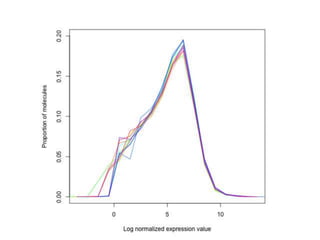
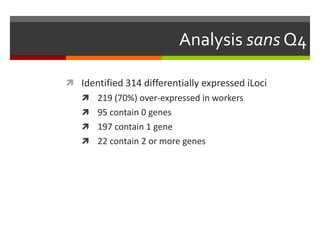
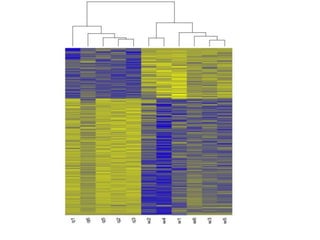
The document summarizes Daniel Standage's analysis of differential gene expression in the paper wasp Polistes dominula. He performed locus-level differential expression analysis after isoform-level analysis proved unreliable. The genome contains 18,675 intervals (iLoci) partitioned by gene content. Initial analysis found 295 differentially expressed iLoci but sample grouping was problematic. After filtering iLoci by read counts and re-running the analysis without a problematic sample, 314 differentially expressed iLoci were identified, mostly over-expressed in workers. Cytochrome P450 and NADH dehydrogenase genes were frequently differentially expressed.