This document describes an automated robotic biorepository system developed by researchers at the University of Virginia to store and retrieve genomic DNA samples on a large scale. The system integrates three primary devices - a robotic arm, liquid handling robot, and microplate storage system. It is capable of automatically processing, analyzing, storing, and retrieving up to 250,000 genomic DNA samples in microplates held at -80°C. The system uses barcoded microplates and tubes to track samples and a database to store associated demographic and analysis data. Its main functions are to create master plates from samples, generate daughter plates for storage and retrieval, and assemble send-out plates of selected samples for distribution.
![A
vailability of the draft ver-
sion of the human genome
provides investigational op-
portunities to comprehen-
sively study new genes and the cor-
responding control of cellular
reactions.1
In order to support these
studies, there will be a parallel need
for purified human genomic DNA
samples from characterized popula-
tions of subjects as starting materi-
als. Moreover, biographical/demo-
graphic information describing each
sample will be essential for the com-
plete understanding of the complex
interactions of the human genome
with its environment. Although these
samples can be obtained using sev-
eral methods, one efficient approach
would be to automate both speci-
men management and collection of
the specimen’s demographic infor-
mation. This streamlined approach
contrasts with the large majority of
biorepositories that manually pro-
cess, store, and retrieve samples and
manually store and retrieve speci-
men information from a database.
The creation of the automated
biorepository can therefore be con-
sidered a direct consequence of the
increased need for specimens and
information in a timely, cost-effec-
tive manner.
Many of the individual laboratory
tasks associated with sample han-
dling, such as analysis, dilution, aspi-
ration and delivery, and archival stor-
age and retrieval, have already been
automated. The challenge is to inte-
grate these laboratory activities into
a seamless operation that delivers
walkaway automation yet permits
user input at key points in the sample
processing cycle. In collaboration
with the North Shore University Hos-
pital and AMDeC (Academic Medical
Development Corp., New York, NY,
www.amdec.org), the Medical Au-
tomation Research Center (MARC)
at the University of Virginia (Char-
FEATURE ARTICLE
Theodore E. Mifflin, B. Sean Graves, and Robin A. Felder
A Large-Scale Robotic Storage and Retrieval
System (Biorepository) Capable of Fully
Automated Operation
Abstract
A robotic system to process, analyze, store, and archivally retrieve genomic samples (DNA and plasma) has been devel-
oped. Its primary function is the automated storage and retrieval of selected samples for distribution according to sam-
ple attributes. The robotic biorepository integrates the operation of three primary devices (track robotic arm, three-axis
pipetting workstation, and large-capacity microplate storage system) into a seamless operation that performs a series of
tasks in a walkaway manner. A user-friendly interface (Visual Basic [Microsoft, Redmond, WA]) allows for operator selec-
tion of tasks and provides for real-time monitoring of events. The system’s operation is synchronized by two PCs, one to
control its tasks, and the other to store demographic and other sample-specific information for later access using Mi-
crosoft SQL. The system is developed around the bar-coded microplate as a common means of storage, analysis, and
distribution with a maximum capacity of approx. 2500 microplates or 250,000 samples. Additional functionality such as
thermal cycling and DNA array spotting, which would be coupled to the biorepository, are modules to be developed.
Handling and storage of other sample types such as RNA, cell suspension, and tissues are also being considered.
Key Words
Biorepository, robotic, automation, liquid handling, cherry picking, microplate, genomic DNA, analysis
The authors are with the University of Virginia, Department of Pathology, Medical Automation Research Center
(MARC), P.O. Box 800214, Charlottesville, VA 22908, U.S.A.; tel.: 804-924-8215; fax: 804-924-5718; e-mail:
tem6h@virginia.edu. The authors wish to recognize the many contributions made by the following members of MARC
during the design, development, and construction of the biorepository: Dr. Glen Wasson, Mr. Jim Gunderson, Mr.
Steve Kell, Ms. Sarah Woods, and Ms. Catherine Piche. Ms. Elle Kovarikova provided the animation and rendered im -
ages used in Figure 1. The authors also wish to recognize the extensive collaboration of persons at the project’s sponsor
(AMDeC): Dr. Peter Gregersen, Mr. Robert Lundsten, and Mr. Houman Khalili. This project would not have been com -
pleted without their efforts. Several hardware vendors are also acknowledged, including TECAN Instruments U.S.
(Durham, NC) and CRS Robotics (Toronto, Canada), for their substantial assistance regarding both the hardware
and software issues that arose during the biorepository’s development.](https://image.slidesharecdn.com/6728b079-6ffe-4e76-8da6-2f59722a9ce5-160808022536/85/biorepository-1-320.jpg)
![BIOREPOSITORY continued
lottesville, VA) has designed and con-
structed a large-scale biorepository
that maximizes the use of robotics
and automation to achieve this objec-
tive (see marc.med.virginia.edu and
Ref. 2). In its current form, the
robotic biorepository has the capac-
ity to automatically accession, dis-
pense, analyze, store, and randomly
access up to 250,000 genomic DNA
samples. Additional capability is now
being developed to manipulate hu-
man blood plasma as well as RNA
and cell suspensions.
Hardware
The core components of the ba-
sic biorepository design are three
automated devices: an articulated
robot arm mounted on a 3-m track
(T265, CRS Robotics, Toronto, Ca-
nada), a three-axis liquid handing
robot (Genesis 150 RSP, TECAN In -
struments U.S., Durham, NC), and
a tiered microplate storage system
(MolBank [GIRA], TECAN Instru -
ments ) capable of holding up to
2500 standard microplates at 4 °C.
Additional functionality is provided
by several secondary devices that
are connected to the primary de-
vices either mechanically or through
electronic means (e.g., serial ports).
These secondary devices include
microplate heat sealer, balance, mi-
croplate chiller, and a microplate
reader capable of quantifying UV
and fluorescence measurements.
They are arranged on the track
robot’s table in a manner that makes
them accessible to the track robot
arm (Figure 1). The track robot has
a five-axis arm with a gripper capa-
ble of holding both tubes and mi-
croplates, depending on the grip
width of the two sets of fingers
attached to the gripper itself. Two
interconnected computers (Dell
Computer , Inc., Austin, TX) are
used for operating the biorepository
and are run on Microsoft Windows
NT. One PC (the controller) orches-
trates the operation of the various
pieces of hardware, while the sec-
ond PC (the database) functions as
a host for the database (Microsoft
server) to store demographic infor-
mation. A series of uninterruptable
power supplies maintain constant
power into the biorepository.
Software
Similar to the hardware, the soft-
ware operating system was designed
using a mixture of application pack-
ages.3
Several of these packages were
provided with the core components,
such as the operating system for the
robotic arm (CROSnt) (CRS Ro -
botics ) and the operating system for
“THE ROBOTIC BIOREPOSI-
TORY INTEGRATES THE
OPERATION OF THREE
PRIMARY DEVICES INTO A
SEAMLESS OPERATION THAT
PERFORMS A SERIES OF
TASKS IN A WALKAWAY
MANNER.
”
Figure 1 Illustration of robotic biorepository. The robotic arm’s 3-m track tra -
verses the length of the table and ends at the MolBank (right side) while the Gen -
esis is positioned so that it can be accessed by the arm (middle of track). Other
accessories are positioned around the table within reach of the track robot. Sam -
ples (in 50-mL tubes) are placed near the MolBank in four racks of 24
samples/rack (foreground of table). Microplates are loaded into the carousel op -
posite the Genesis (far corner) next to the microplate sealer.
Figure 2 Software organization on the robotic biorepository. The user interface
and the CROS system software reside on the controller PC, whereas the SQL re -
sides on the server PC. The controller PC communicates with most of the acces -
sory devices via their serial ports, while the MolBank is connected via a custom-
written interface protocol. The arrows show the data flow direction.](https://image.slidesharecdn.com/6728b079-6ffe-4e76-8da6-2f59722a9ce5-160808022536/85/biorepository-2-320.jpg)
![the Genesis pipetting station (Gem-
ini). Connectivity to the remaining ac-
cessories was accomplished using se-
rial ports on the accessories and
coupled to the operational PC via a
single communication system (Cy-
clades-Z, Cycleades Corp., Fre-
mont, CA). The entire biorepository
is interconnected using a network
based on POLARA™ (CRS Robot -
ics ). The biorepository’s operation is
normally controlled using a custom-
written user interface (UI) and oper-
ating system based on Visual Basic
(VB) and RAPL-3 (Robotic Application
Programming Language-3, CRS
Robotics ) (Figure 2), but may also be
controlled via an Internet connection
using PC anywhere. In addition to an
integrated operation mode, several of
the accessory devices (e.g., Spec-
traFluor Plus microplate analyzer
[TECAN-US, Research Triangle Park,
NC] and Sartorius [Edgewood, NJ]
balance) can be operated in an inde-
pendent mode. This standalone opera-
tion offers flexibility for the bioreposi-
tory operators to develop other
applications, while using existing
hardware resources more efficiently.
Consumables
The robotic biorepository uses
five different disposables for pro-
cessing, analyzing, storing, and re-
trieving genomic DNA (Figure 3).
Their characteristics (summarized in
Table 1) illustrate the diverse prop-
erties needed for the successful op-
eration of the biorepository. For ex-
ample, the storage microplates
(items 1, 2, and 5) are constructed
from polypropylene to withstand
low temperatures (to –80 °C) and are
sterile to prevent degradation of ge-
nomic DNA from stray nucleases.
For the protein-based human
plasma, a sixth type of storage sys-
tem has been incorporated that uses
a two-dimensional matrix code
(TrakMate™, Matrix Corp., Hud-
son, NH) on individual tubes (racks
of 96 matrix-coded plasma tubes are
also liner bar-coded). Individual mi-
croplates (and brands) are selected
for the attributes that best match
their role in the biorepository’s oper-
ation (Table 1). All consumables
contain linear bar codes for perma-
nent identification.
Biorepository operation
summary
The robotic biorepository is an in-
tegrated system that consists of a
group of hardware items, software
packages, and procedures that per-
form a concise menu of tasks. The
biorepository stores genomic DNA in
a compacted format with the major-
ity (>98%) of each sample frozen at
–80 °C. Most (~80%) of the genomic
DNA is contained in five deep-well
master plates (Table 1), while a
lesser fraction (2–18%) resides frozen
in up to nine daughter plates (Table
1). Only about 2% of the genomic
DNA is immediately available for
continual access (one daughter
plate) at 4 °C. Sendout plates contain
the collection of specimens identi-
fied from a search of the database for
specimens that match particular cri-
teria. Aliquots from these specimens
are removed from selected daughter
plates and are delivered into the
sendout plate for delivery to an in-
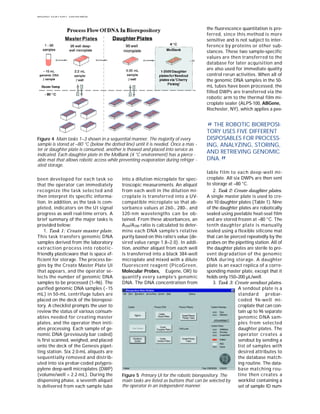
vestigator (Figure 4).
There are four main tasks of the
biorepository: 1) Create master
plates, 2) create daughter plates, 3)
create sendout plates, and 4) create
master plasma tubes. Each of these
tasks can be selected separately
from the primary UI (Figure 5). Ad-
ditional custom-designed UIs have
“THE DNA CONCENTRA-
TION FROM THE FLUO-
RESCENCE QUANTITATION
IS PREFERRED, SINCE THIS
METHOD IS MORE SENSITIVE
AND IS NOT SUBJECT TO
INTERFERENCE BY PROTEINS
OR OTHER SUBSTANCES.
”
Figure 3 Six disposable (sterile) microplates used for the robotic biorepository.
Shown counterclockwise from lower left are: 1) master plate, 2) daughter plate
with pierceable mat, 3) UV spectra microplate, 4) black (fluorescent quantita -
tion) microplate, 5) sendout plate sealed with peelable film, and 6) rack of
plasma tubes with 2-D matrix-coded tubes.
Table 1
Biorepository consumable items and their key attributes
Item Material No. of Sterile Storage Sealed
no. Item stored wells (Y/N) Composition temp. (Y/N)
1 Master plate Genomic DNA 96 Y Polypropylene –80 °C Y
2 Daughter plate Genomic DNA 96 Y Polypropylene –80 °C Y
3 UV analysis None 96 N Polystyrene/? — N
4 Fluor. quant. None 384 N Polystyrene — N
5 Sendout plate Genomic DNA 95 Y Polypropylene to Y
–80 °C
6 Plasma master Plasma 96 Y Polypropylene –80 °C *
tubes
*Tubes are sealed individually using sterile strip caps](https://image.slidesharecdn.com/6728b079-6ffe-4e76-8da6-2f59722a9ce5-160808022536/85/biorepository-3-320.jpg)